Hi sen2cor processing show me this error ans it repeats for each granule .
i use both snap and command window to launch the processing and i alwyad get the same error:
any help please and thanks in advance
Hi sen2cor processing show me this error ans it repeats for each granule .
i use both snap and command window to launch the processing and i alwyad get the same error:
This validation problem is a warning and should not affect your results. It is due to some product xml file not complying to xsd (xml spec.) of the sen2cor.
it affects my resulats. indeed when i open the product i do not find the bands as you see here:
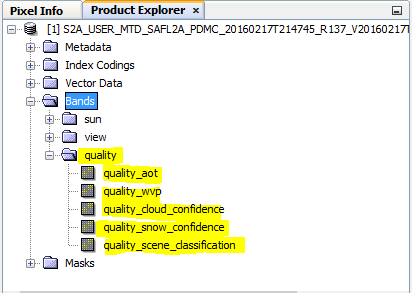
error.txt (75.1 KB)
that’s the error message
Are you using the 60m reader? Your product is processed at 60m so it only has 60 bands, using a 10m or 20m reader could lead to your result.
i did not understand what youmean by reader can you explain more please?
i tried it but i got the same result
Could you try to open a jp2 file in GRANULE/[granule name]/IMG_DATA? Could you try to open a single granule in GRANULE/[granule name], still with the 60m reader (other reader won’t show bands)? Thanks.
You can’t open or process a granule with Sen2Cor, but you should be able to open the product or a granule with SNAP with the 60m reader.
the granule not processed yet with sen2cor it’s like that:
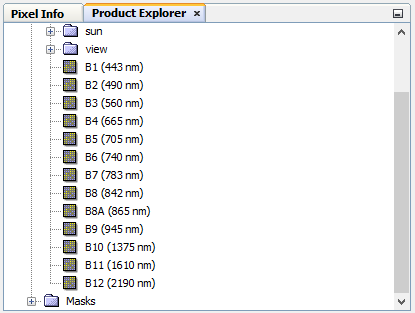
it has all bands but when i process the image i get this:
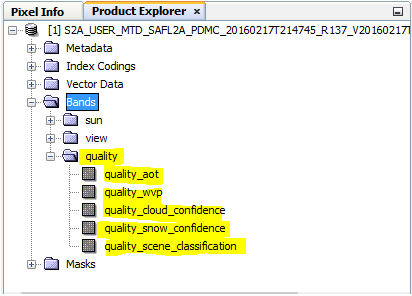
do you have any idea about what should i do ? thank you
Under the granule folder, you should have a “IMG_DATA/60m” folder with 14 jp2 files in it. Can you check this and could you try to open these files in SNAP?
As well, when I said to use the 60m reader I meant when you open the L2A product in SNAP a dialog appears where you can select the reader. You have to select “Sentinel-2 MSI L2A - resampled at 60m resolution - WGS84 / UTM zone XXX”. Was it clear on my previous messages?
i did just what you told me but i get the same results . here’s the folder you said it will contains14 jp2
This means SNAP did ok, the problem s related to Sen2Cor and I don’t know what the problem could be. @umwilm should be able to help you with this.
thank you so much i really appreciate it
The way how you called the processor (with giving the complete path up to the img_data folder) has probably created a corrupted L2A product (see that it tries to create a datastrip folder) inside the img_data folder. Remove the L2A product and start again, hereby only giving the name of the L1C product folder as an argument for your processing.
this is only a warning that the metadata did not pass the validation. The reasons are given in the logfile. It does not affect the image generation itself.