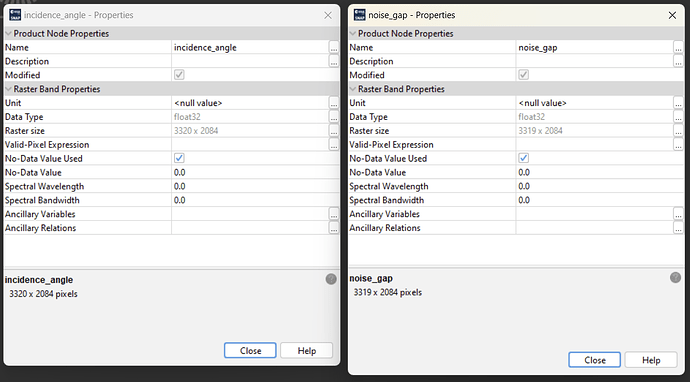
I’ve encountered mismatching raster dimensions after slice assembling and multi-looking Sentinel-1 GRD acquisitions.
Starting with the three products
S1A_IW_GRDH_1SDV_20250416T154949_20250416T155014_058784_074866_7CDF.SAFE
S1A_IW_GRDH_1SDV_20250416T155014_20250416T155039_058784_074866_4586.SAFE
S1A_IW_GRDH_1SDV_20250416T155039_20250416T155104_058784_074866_23CC.SAFE
I applied the operators Calibration and ThermalNoiseRemoval on each using SNAP GPT and then applied SliceAssembly, Multilook with a factor of 8 and finally LinearToFromdB.
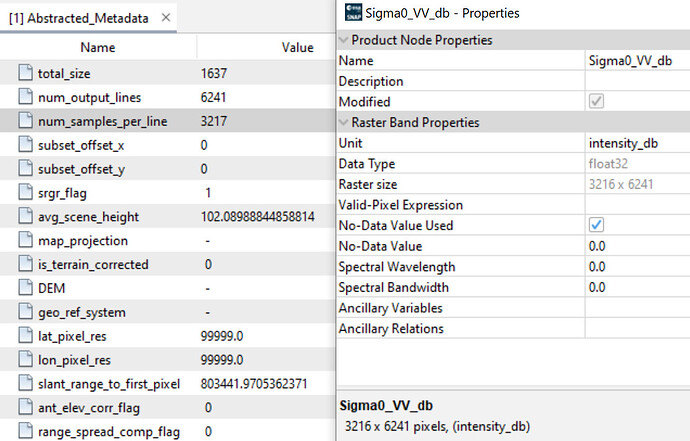
In the resulting BEAM-DIMAP file S1A_IW_GRDH_1SDV_20250416T154949_20250416T155104_058784_074866_7CDF_Cal_tnr_Asm_ML_dB.dim there are mismatching values for the raster width:
...
<Raster_Dimensions>
<NCOLS>3217</NCOLS>
<NROWS>6241</NROWS>
<NBANDS>2</NBANDS>
</Raster_Dimensions>
...
<Image_Interpretation>
<Spectral_Band_Info>
<BAND_INDEX>0</BAND_INDEX>
<BAND_DESCRIPTION />
<BAND_NAME>Sigma0_VV_db</BAND_NAME>
<BAND_RASTER_WIDTH>3216</BAND_RASTER_WIDTH>
<BAND_RASTER_HEIGHT>6241</BAND_RASTER_HEIGHT>
...
</Spectral_Band_Info>
<Spectral_Band_Info>
<BAND_INDEX>1</BAND_INDEX>
<BAND_DESCRIPTION />
<BAND_NAME>Sigma0_VH_db</BAND_NAME>
<BAND_RASTER_WIDTH>3216</BAND_RASTER_WIDTH>
<BAND_RASTER_HEIGHT>6241</BAND_RASTER_HEIGHT>
...
</Spectral_Band_Info>
So the whole product is supposed to have 3217 columns, but the bands both actually only have a width of 3216 pixels.
In SNAP, this is also shown accordingly:
This tripped the BEAM-DIMAP reader I wrote for further processing SNAP data in Python using xarray and I guess it is an error somewhere along the processing chain and shouldn’t happen.