The command-line often makes it easier to discover the reason something doesn’t work, and you can paste details into a post so it is easy for others to reproduce the problem. I’m not sure if you are doing standard C2RCC processing or have some special requirements, but it is often helpful to start with a basic example following the SNAP Help pages.
I was able to process A2009246231500.L1A_LAC (the file used for the examples in the SNAP Help) using the current OCSSW software and following the Help documentation.
Note that the % is my zsh prompt – you may be using bash which will give a $ prompt:
% modis_GEO -v A2009246231500.L1A_LAC
** Files utcpole.dat/leapsec.dat are more than 2 weeks old.
** Running update_luts.py to update files...
[...]
eogen_modis returned with exit status: 0
Percentage of pixels with missing geolocation: 0.00
Validation test passed for geolocation file A2009246231500.GEO
geogen_modis created A2009246231500.GEO successfully!
MODIS geolocation processing complete.
% modis_L1B A2009246231500.L1A_LAC
MODIS Aqua L1B version 6.2.1_obpg, built Jan 26 2021 14:56:49
scan: 0 out of 203 Wed Apr 21 14:50:07 2021
[...]
scan: 200 out of 203 Wed Apr 21 14:50:18 2021
gwhite@cerberus ~/Documents/ESA_SNAP/C2RCC
% l2gen par=L1C.par
Input file A2009246231500.L1B_LAC is MODIS Aqua Level-1B HDF-EOS product.
[...]
Percentage of pixels flagged:
Flag # 1: ATMFAIL 0 0.0000
Flag # 2: LAND 3006 0.1094
Flag # 3: PRODWARN 0 0.0000
Flag # 4: HIGLINT 540178 19.6527
Flag # 5: HILT 1130435 41.1274
Flag # 6: HISATZEN 188459 6.8565
Flag # 7: COASTZ 486 0.0177
Flag # 8: SPARE 0 0.0000
Flag # 9: STRAYLIGHT 950625 34.5855
Flag #10: CLDICE 1553519 56.5200
Flag #11: COCCOLITH 0 0.0000
Flag #12: TURBIDW 0 0.0000
Flag #13: HISOLZEN 0 0.0000
Flag #14: SPARE 0 0.0000
Flag #15: LOWLW 0 0.0000
Flag #16: CHLFAIL 0 0.0000
Flag #17: NAVWARN 0 0.0000
Flag #18: ABSAER 0 0.0000
Flag #19: SPARE 0 0.0000
Flag #20: MAXAERITER 0 0.0000
Flag #21: MODGLINT 1116643 40.6256
Flag #22: CHLWARN 0 0.0000
Flag #23: ATMWARN 0 0.0000
Flag #24: SPARE 0 0.0000
Flag #25: SEAICE 0 0.0000
Flag #26: NAVFAIL 0 0.0000
Flag #27: FILTER 0 0.0000
Flag #28: SPARE 0 0.0000
Flag #29: BOWTIEDEL 0 0.0000
Flag #30: HIPOL 144971 5.2743
Flag #31: PRODFAIL 0 0.0000
Flag #32: SPARE 0 0.0000
End MSl12 processing at 2021111150623000
Processing Rate = 21.827957 scans/sec
Processing Completed
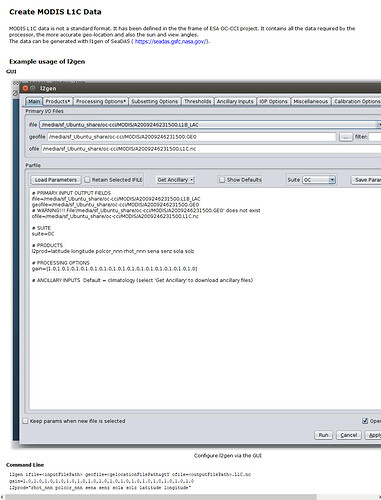
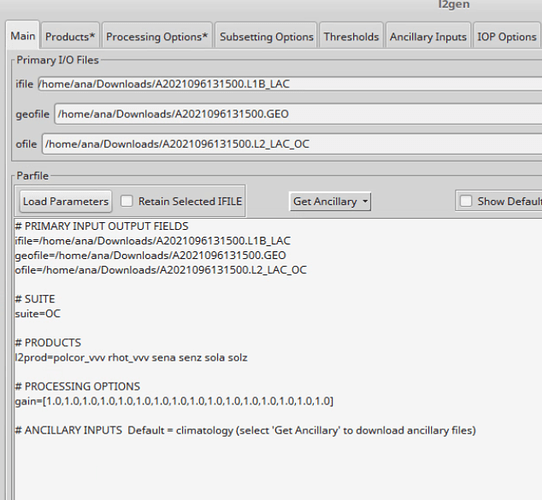
The par file was copied from the Help pages:
% cat L1C.par
ifile=A2009246231500.L1B_LAC
geofile=A2009246231500.GEO
ofile=A2009246231500.L1C.nc
suite=OC
l2prod=latitude longitude polcor_nnn rhot_nnn sena senz sola solz
gain=[1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0,1.0]
atmocor=off
I was able to use the GUI default setting to get A2009246231500.L1C.nc_C2RCC.dim. The metadata
has:
<PRODUCT_SCENE_RASTER_START_TIME>03-SEP-2009 23:20:00.253000</PRODUCT
_SCENE_RASTER_START_TIME>
<PRODUCT_SCENE_RASTER_STOP_TIME>03-SEP-2009 23:15:01.877000</PRODUCT_
SCENE_RASTER_STOP_TIME>
[...]
<MDATTR name="time_coverage_start" type="ascii">2009-09-03T23:15:01.877Z</MDATTR>
<MDATTR name="time_coverage_end" type="ascii">2009-09-03T23:20:00.253Z</MDATTR>
I also ran with the parameters changed to include the angles:
<parameters>
<validPixelExpression>!(l2_flags.LAND || max(rhot_412,max(rhot_443,max(rhot_488,max(rhot_531,max(rhot_547,max(rhot_555,max(rhot_667,max(rhot_678,max(rhot_748,rhot_869)))))))))>0.25)</validPixelExpression>
<salinity>35.0</salinity>
<temperature>15.0</temperature>
<ozone>330.0</ozone>
<press>1000.0</press>
<outputRtosa>false</outputRtosa>
<outputAsRrs>false</outputAsRrs>
<outputAngles>true</outputAngles>
</parameters>
On the command line (using default output:target.dim, the % character is the zsh prompt; bash uses $):
% gpt C2RCC -e -SsourceProduct=A2009246231500.L1C.nc \
-PsensorName=modis \
-PvalidPixelExpression='!(l2_flags.LAND || max(rhot_412,max(rhot_443,max(rhot_488,max(rhot_531,max(rhot_547,max(rhot_555,max(rhot_667,max(rhot_678,max(rhot_748,rhot_869)))))))))>0.25)' \
-Psalinity=35.0 \
-Ptemperature=15.0 \
-Pozone=330.0 \
-Ppress=1000.0
INFO: org.esa.snap.c\
ore.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Incompatible GDAL 3.1.4 found on system. Internal GDAL 3.0.0 from distribution will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.
INFO: org.hsqldb.persist.Logger: dataFileCache open start
Executing operator...
20%....30%....40%....50%....60%....70%....80%....90%.... done.
INFO: org.esa.snap.core.gpf.common.WriteOp: Start writing product c2rcc.modis$178F64B7F0E to target.dim
Writing...
10%INFO: org.esa.s3tbx.c2rcc.ancillary.AtmosphericAuxdataBuilder: Atmospheric auxdata product can't be used. At least one is not specified. Using constant values for ozone (330.0) and surface pressure (1000.0).
20%40%....50%....60%....70%....80%....90%....100%....110%.... done.
INFO: org.esa.snap.core.gpf.common.WriteOp: End writing product target to target.dim
INFO: org.esa.snap.core.gpf.common.WriteOp: Time: 18.076 s total, 8.904 ms per line, 0.006576 ms per pixel