Good evening all!
I’m a new user of snap2stamps, so I already thank in advance whoever will try to help me.
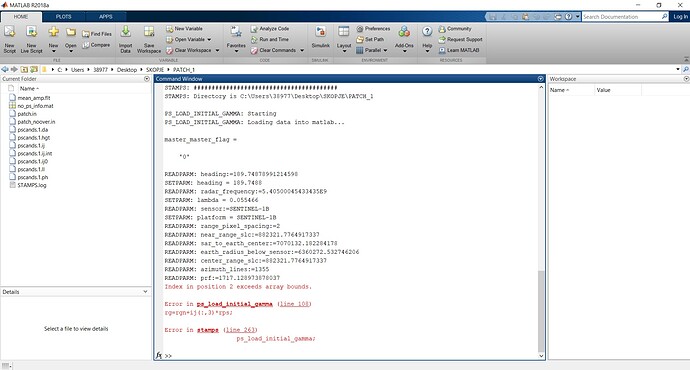
The reason for my new topic is that I’m having an error when I run the mt_snap_prep script.
What I have done until now is (after obviously have done every other previous step smoothly):
- I created a new folder “stamps” in the project folder;
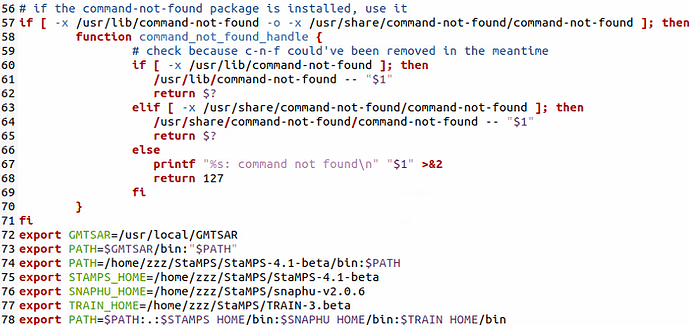
- in this folder I call the terminal and I write source /…/StaMPS_CONFIG.bash (after having modified the script with the correct dir of StaMPs 4.1b, Triangle (bin) and Snaphu (bin) and the addition of a last line exportPATH=${PATH}:$STAMPS/bin:$MATLABPATH);
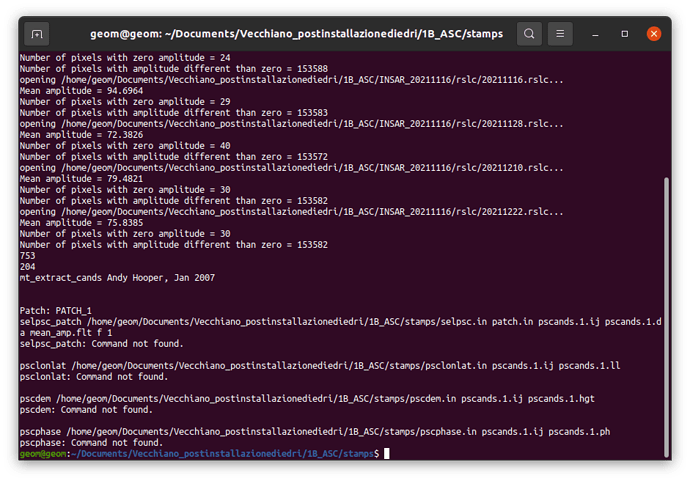
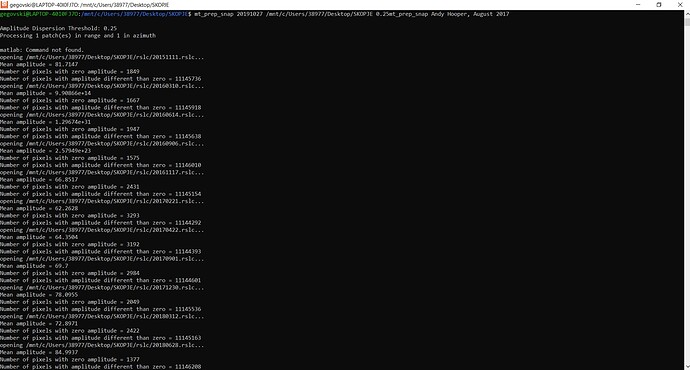
- If I call mt_prep_snap master_date /…/INSAR_master_date/ 0.42 is giving me mt_prep_snap command not found, I tried also removing the last / from /…/INSAR_master_date, again not working;
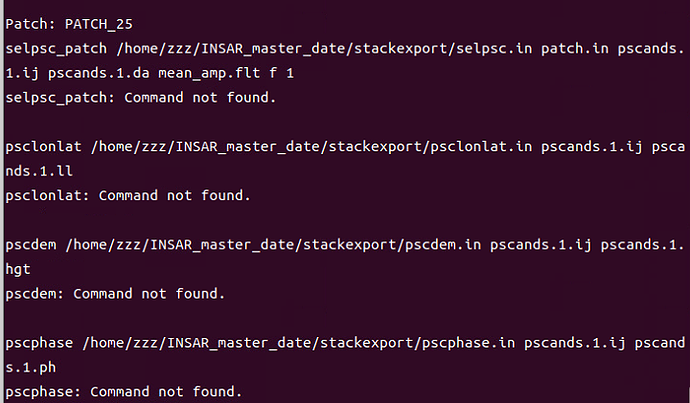
- similarly to the previous steps, I tried with copying in the new stamps folder all the files present in the bin folder of StaMPs 4.1b and if I call mt_prep_snap master_date /…/INSAR_master_date/ 0.42 is working…but is giving the errors shown in the screenshots.
Somehow it does not find the files selpsc_path, psclonlat, pscdem and pscphase commands.
What I am doing wrong?
I followed the workflow as showed but I don’t know why is giving me this.
I have respectively 6 (1B-Asc) and 8 (1A-Asc) slave images (I need to check if the artificial permanent scatterers we have installed are visible, so I am considering only the images from the 12/10/2021 until now).
Additional info:
“StaMPS should be compiled in gcc-7” - I checked and is gcc-7 selected with the *.
I don’t have the Segmentation Fault (core dumped) error.
Interferograms after the coreg_ifg_topsar.py seem normal, also the intensity images.
Thanks again in advance!
Hope to find a solution!