Hi everyone,
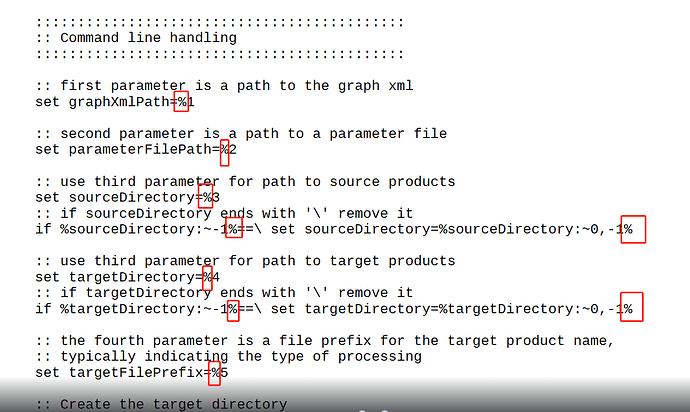
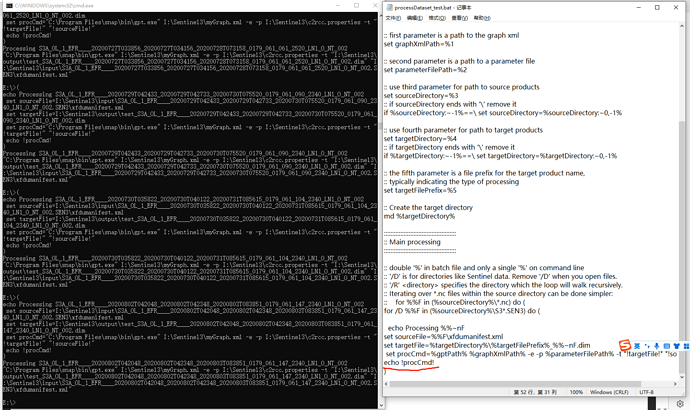
I am having trouble running C2RCC on OLCI. I have set up a processDataset.bat file and a c2rcc_olci_proc xml file (see attached).
when I run the commend
.\processDataset.bat .\c2rcc_olci_proc.xml "H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\" "H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\" C2RCC
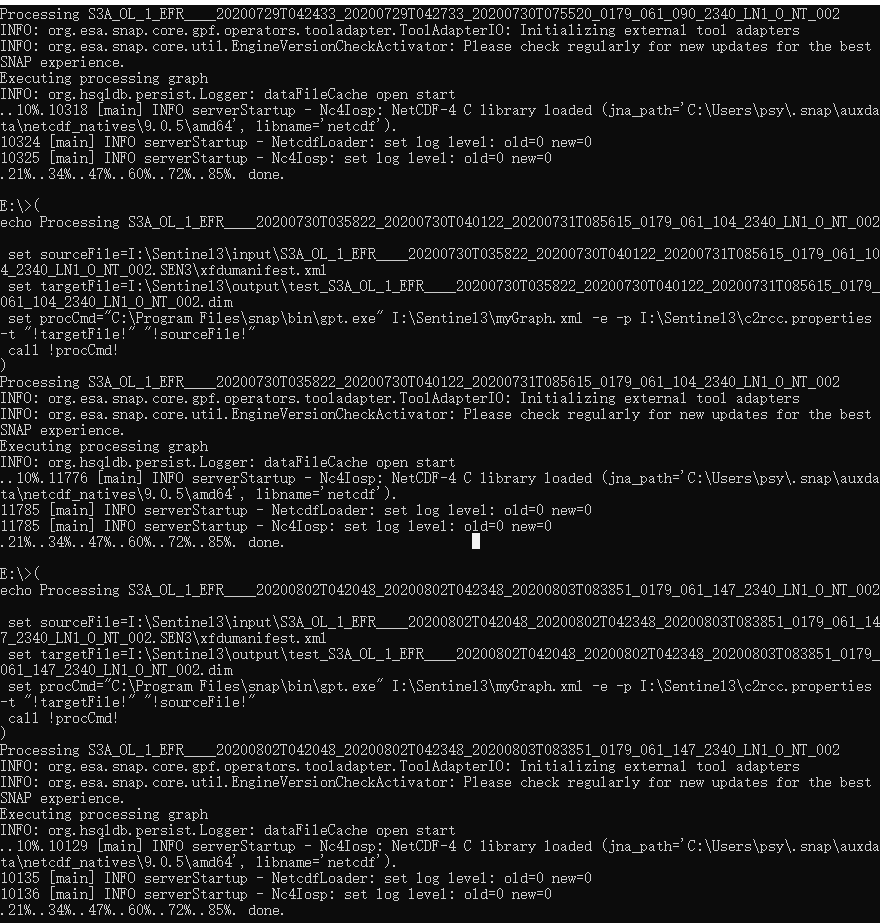
it loops over the files I put in a test folder (2 files) but for each file I get an Error: -t (The system cannot find the file specified).
I am not sure why this is happing as the file paths seem to be correct. I tired pointing to the S3 folder itself as well as the xfdumanifest.xml file but get the same error. I tried looking through the forum but couldn’t find a similar issue.
Thank you for your help!
here is the full output from the run:
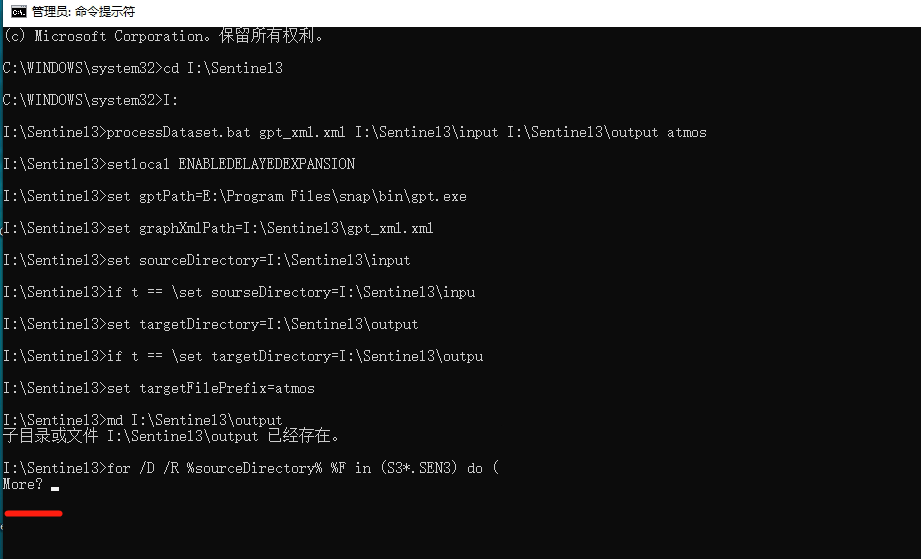
(satenv) PS C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP> .\processDataset.bat .\c2rcc_olci_proc.xml "H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\" "H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\" C2RCC
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>setlocal ENABLEDELAYEDEXPANSION
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>set gptPath="C:\Users\bol_remote_sensing\AppData\Local\Programs\snap\bin\gpt.exe"
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>set graphXmlPath=.\c2rcc_olci_proc.xml
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>set sourceDirectory=H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>if \ == \ set sourceDirectory=H:\BOL_satellite_data\Arctic\YK_S3\level_1\test
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>set targetDirectory=H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>if \ == \ set targetDirectory=H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>set targetFileSuffix=C2RCC
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>md H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017
A subdirectory or file H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017 already exists.
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>for / %F in (S3*_x) do (
echo.
set sourceFile=%~fF\xfdumanifest.xml
echo Processing !sourceFile!
set targetFile=H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\%~nF_C2RCC.nc
set procCmd="C:\Users\bol_remote_sensing\AppData\Local\Programs\snap\bin\gpt.exe" .\c2rcc_olci_proc.xml -e -p -t "!targetFile!" "!sourceFile!" -f NetCDF4-CF
call !procCmd!
)
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>(
echo.
set sourceFile=H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\S3A_OL_1_EFR____20171030T225128_20171030T225253_20180721T045623_0085_024_044_1800_LR2_R_NT_002_x\xfdumanifest.xml
echo Processing !sourceFile!
set targetFile=H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\S3A_OL_1_EFR____20171030T225128_20171030T225253_20180721T045623_0085_024_044_1800_LR2_R_NT_002_x_C2RCC.nc
set procCmd="C:\Users\bol_remote_sensing\AppData\Local\Programs\snap\bin\gpt.exe" .\c2rcc_olci_proc.xml -e -p -t "!targetFile!" "!sourceFile!" -f NetCDF4-CF
call !procCmd!
)
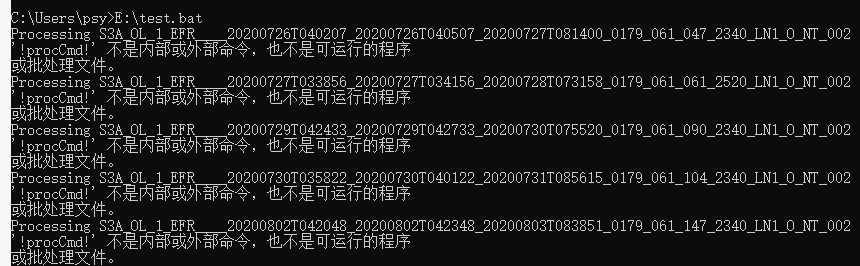
Processing H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\S3A_OL_1_EFR____20171030T225128_20171030T225253_20180721T045623_0085_024_044_1800_LR2_R_NT_002_x\xfdumanifest.xml
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 3.2.1 found on system. JNI driver will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 3.2.1 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
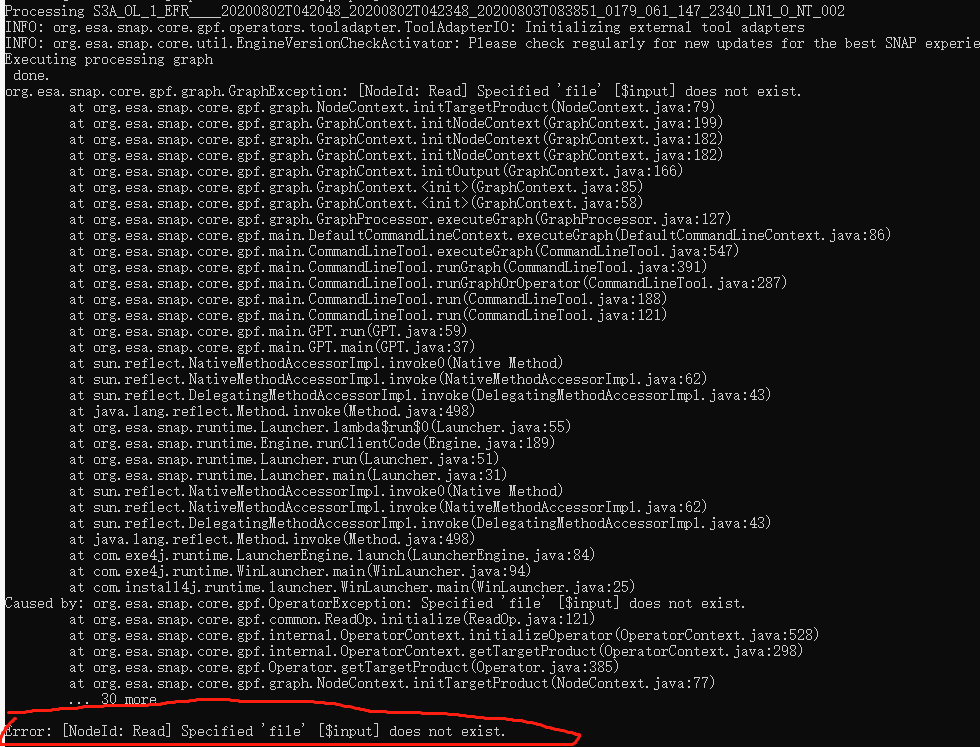
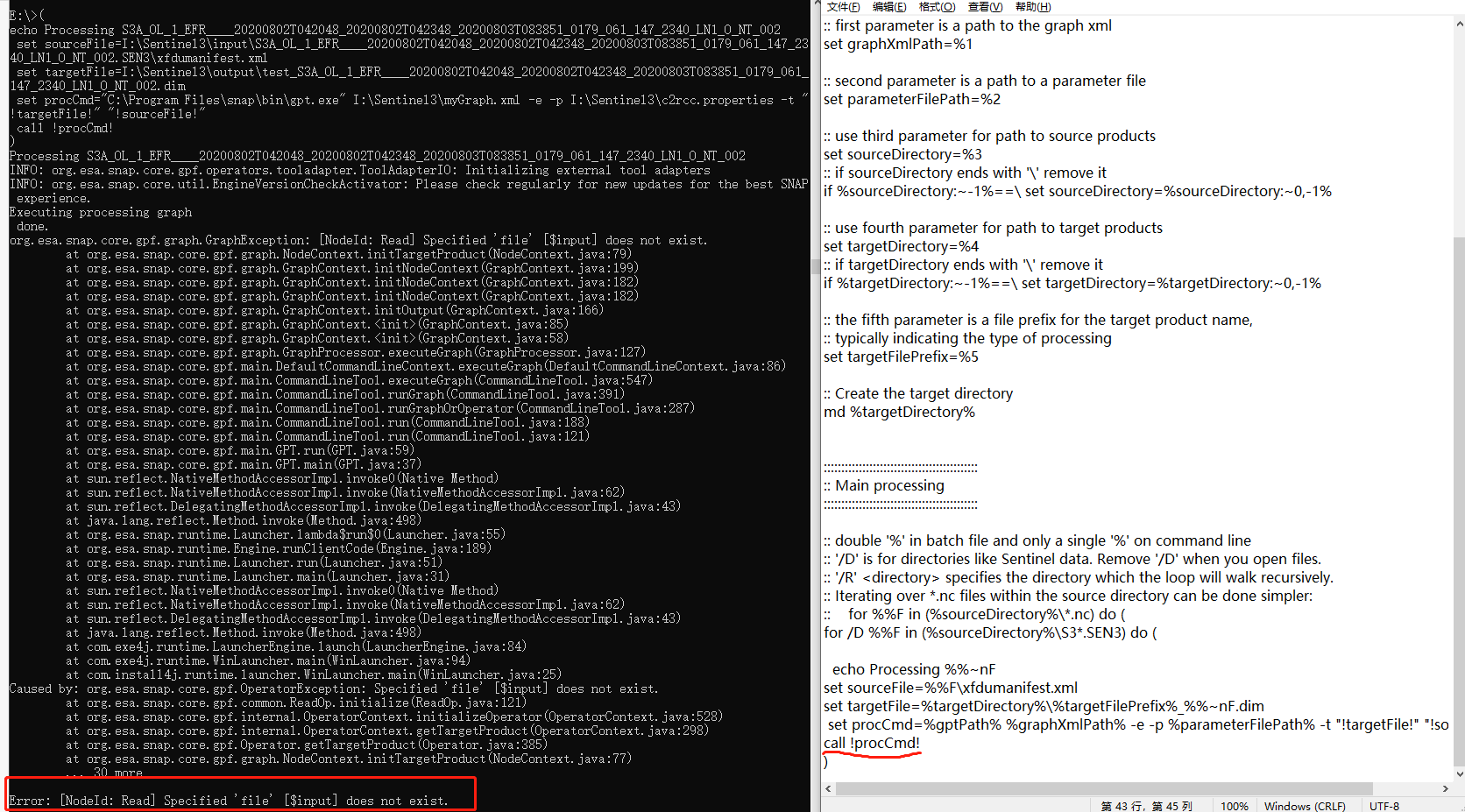
java.io.FileNotFoundException: -t (The system cannot find the file specified)
at java.io.FileInputStream.open0(Native Method)
at java.io.FileInputStream.open(FileInputStream.java:195)
at java.io.FileInputStream.<init>(FileInputStream.java:138)
at java.io.FileInputStream.<init>(FileInputStream.java:93)
at java.io.FileReader.<init>(FileReader.java:58)
at org.esa.snap.core.gpf.main.DefaultCommandLineContext.createReader(DefaultCommandLineContext.java:101)
at com.bc.ceres.metadata.MetadataResourceEngine.readResource(MetadataResourceEngine.java:94)
at org.esa.snap.core.gpf.main.CommandLineTool.getRawParameterMap(CommandLineTool.java:498)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraph(CommandLineTool.java:334)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraphOrOperator(CommandLineTool.java:287)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:188)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:121)
at org.esa.snap.core.gpf.main.GPT.run(GPT.java:59)
at org.esa.snap.core.gpf.main.GPT.main(GPT.java:37)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.esa.snap.runtime.Launcher.lambda$run$0(Launcher.java:55)
at org.esa.snap.runtime.Engine.runClientCode(Engine.java:189)
at org.esa.snap.runtime.Launcher.run(Launcher.java:51)
at org.esa.snap.runtime.Launcher.main(Launcher.java:31)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at com.exe4j.runtime.LauncherEngine.launch(LauncherEngine.java:84)
at com.exe4j.runtime.WinLauncher.main(WinLauncher.java:94)
at com.install4j.runtime.launcher.WinLauncher.main(WinLauncher.java:25)
Error: -t (The system cannot find the file specified)
C:\Users\bol_remote_sensing\remote_sensing\sherman\code\SNAP>(
echo.
set sourceFile=H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\S3A_OL_1_EFR____20171031T204543_20171031T204843_20171102T014240_0180_024_057_1979_LN1_O_NT_002_x\xfdumanifest.xml
echo Processing !sourceFile!
set targetFile=H:\BOL_satellite_data\Arctic\YK_S3\level_2\C2RCC\2017\S3A_OL_1_EFR____20171031T204543_20171031T204843_20171102T014240_0180_024_057_1979_LN1_O_NT_002_x_C2RCC.nc
set procCmd="C:\Users\bol_remote_sensing\AppData\Local\Programs\snap\bin\gpt.exe" .\c2rcc_olci_proc.xml -e -p -t "!targetFile!" "!sourceFile!" -f NetCDF4-CF
call !procCmd!
)
Processing H:\BOL_satellite_data\Arctic\YK_S3\level_1\test\S3A_OL_1_EFR____20171031T204543_20171031T204843_20171102T014240_0180_024_057_1979_LN1_O_NT_002_x\xfdumanifest.xml
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 3.2.1 found on system. JNI driver will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 3.2.1 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
java.io.FileNotFoundException: -t (The system cannot find the file specified)
at java.io.FileInputStream.open0(Native Method)
at java.io.FileInputStream.open(FileInputStream.java:195)
at java.io.FileInputStream.<init>(FileInputStream.java:138)
at java.io.FileInputStream.<init>(FileInputStream.java:93)
at java.io.FileReader.<init>(FileReader.java:58)
at org.esa.snap.core.gpf.main.DefaultCommandLineContext.createReader(DefaultCommandLineContext.java:101)
at com.bc.ceres.metadata.MetadataResourceEngine.readResource(MetadataResourceEngine.java:94)
at org.esa.snap.core.gpf.main.CommandLineTool.getRawParameterMap(CommandLineTool.java:498)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraph(CommandLineTool.java:334)
at org.esa.snap.core.gpf.main.CommandLineTool.runGraphOrOperator(CommandLineTool.java:287)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:188)
at org.esa.snap.core.gpf.main.CommandLineTool.run(CommandLineTool.java:121)
at org.esa.snap.core.gpf.main.GPT.run(GPT.java:59)
at org.esa.snap.core.gpf.main.GPT.main(GPT.java:37)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at org.esa.snap.runtime.Launcher.lambda$run$0(Launcher.java:55)
at org.esa.snap.runtime.Engine.runClientCode(Engine.java:189)
at org.esa.snap.runtime.Launcher.run(Launcher.java:51)
at org.esa.snap.runtime.Launcher.main(Launcher.java:31)
at sun.reflect.NativeMethodAccessorImpl.invoke0(Native Method)
at sun.reflect.NativeMethodAccessorImpl.invoke(NativeMethodAccessorImpl.java:62)
at sun.reflect.DelegatingMethodAccessorImpl.invoke(DelegatingMethodAccessorImpl.java:43)
at java.lang.reflect.Method.invoke(Method.java:498)
at com.exe4j.runtime.LauncherEngine.launch(LauncherEngine.java:84)
at com.exe4j.runtime.WinLauncher.main(WinLauncher.java:94)
at com.install4j.runtime.launcher.WinLauncher.main(WinLauncher.java:25)
Error: -t (The system cannot find the file specified)
c2rcc__olci_proc.xml (1.6 KB)
processDataset.bat (1.8 KB)