I have processed 56 Sentinel 1 data with sbas_snap2stamps and it worked fine, but there are some things to note:
-
This is different from snap2stamps which separate slave and master SLC data folders, sbas_snap2stamp combines master and slave file folders.
-
in the configure file, the AOI clip area must be filled in properly, the size must not be more than AOI BBOX DEFINITION.
-
The order of work:
- splitting_master_free
- splitting_slave_free
- sbas_topsar_coreg
- sbas_topsar_ifg
- sbas_export
check the baseline with plotbaseline.py, if something is not connected, you can add it to the sbas_add file, then run sbas_topsar_ifg by taking longer temp_baseline on the configure file.
-
When setting mt_prep_snap rg_patch and az_ptch, for example:
mt_prep_snap 20150119 / min / g / process / stamps / bandung_basin / 149_615 / SBAS / INSAR_20150119 / SMALL_BASELINES / 20141108_20151103 / 0.6 5 5 50 200this is because all data can be stored and processed.
-
This is most important, if the stamps are downloaded from https://github.com/dbekaert/StaMPS/ when the sb_parm_initial command is activated or small_baseline_flag: ‘y’ in stamps (1), stamps step 1 will occur an error.
download and replace the sb_load_initial_gamma.m file from https://github.com/limingjia92/StaMPS/blob/patch-2/matlab/sb_load_initial_gamma.m from the matlab folder.
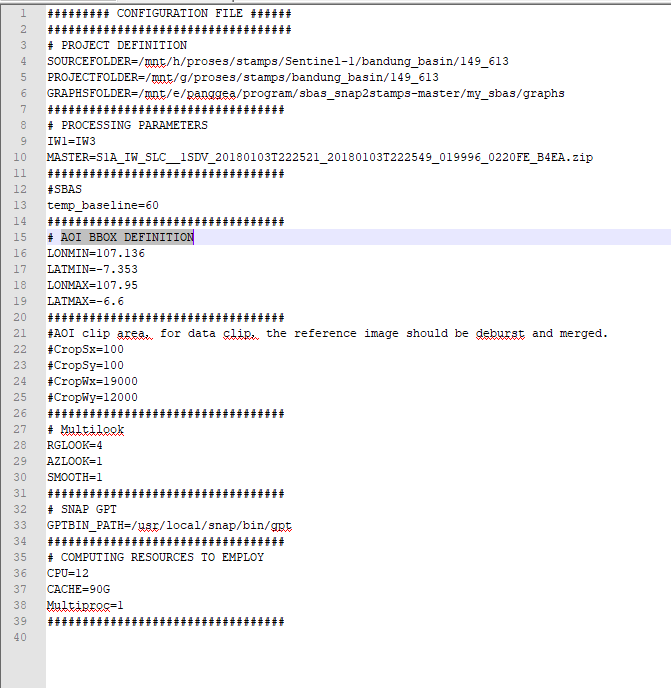
configure file
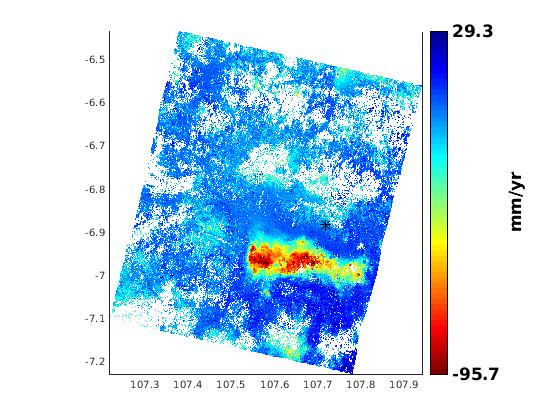
sbas result
.
.
.
and I already process with ps insar
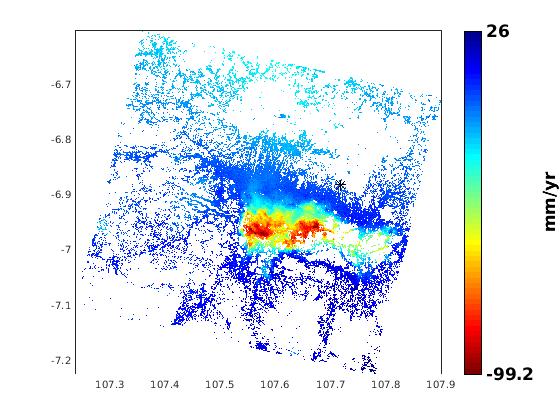
ps result