Hi @dongyusen
Thanks for sharing with us SBAS to StaMPS
I would like to test , so that I have this Error in the first step of splitting_master_free. Please have look to my print Screen to fix my problem.
.
Cheers.
Hi @dongyusen
Thanks for sharing with us SBAS to StaMPS
I would like to test , so that I have this Error in the first step of splitting_master_free. Please have look to my print Screen to fix my problem.
.
Cheers.
Dear Dongyusen,
Please provide a clear processing steps regarding your code on SBAS_SNAP2StaMPS.
Thank you so much.
Shouldn’t LATMIN be smaller than LATMAX?
Do you want to say that my data folder include all slaves and the master image??
Sorry, but this is not my txt, mine is here
the master splitting script work fine,but I got an error when I run the slave_splitting_free.py
the error continued appearting and I had to close the powershell window to shutdown the program. Can you help me?
Thank you in advance
Hi @Linya-Peng ,
I want to test SBAS_snap2stamps , and I got the same error when splitting slaves,
I read your configuration file. Does your folder testdata include all images (slaves and master)??, and what about the folder testinsar ??
Cheers.
Hi @CDE!
Yes, my folder testdata includes all images, both slaves and master. The folder testinsar is the project folder in which log file and result will be. By the way, which OS and python version did you use? I’m a windows user, and I’m not sure whether the error is caused by OS or python version.
Hi @CDE! Me again, I run the script in Ubuntu, and It seemed work well. My python version is python3.8. So I think the wired error was caused by OS.
Hope that can help you~ 
Hi @Linya-Peng , I’m a windows user too. My python version is 2.7
I will try the script in Ubuntu with python3.8 . Thanks for information 
Hi @Linya-Peng @dongyusen ,
which version of snap I must use in Ubuntu ??
when executing : sbas_topsar_ifg.py
I get this error :
Traceback (most recent call last):
File “sbas_topsar_ifg.py”, line 16, in
import numpy as np
ModuleNotFoundError: No module named ‘numpy’
Have you got an idea ??.
Thanks
numpy is a python package which is required to execute the script. You can install it afterwards: NumPy - Installing NumPy
Hi CDE, I’m now using snap8.0 in Ubuntu
Hi @Linya-Peng, @dongyusen, @ABraun
when I execute the command : sbas_topsar_coreg.py
I get this error :
*****IW2 of 1IWs :20161220_20151120 coregistration
b’INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 2.4.2 found on system. JNI driver will be used.’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.4.2 set to be used by SNAP.’
b’INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.’
b’INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.4.2 set to be used by SNAP.’
b’Executing processing graph’
b’INFO: org.hsqldb.persist.Logger: dataFileCache open start’
b"-- org.jblas ERROR Couldn’t load copied link file: java.lang.UnsatisfiedLinkError: /tmp/jblas2130262475707773817/libjblas.so: libgfortran.so.5: Ne peut ouvrir le fichier d’objet partag\xc3\xa9: Aucun fichier ou dossier de ce type."
b’’
b’org.jblas.NativeBlas.dgemm(CCIIID[DII[DIID[DII)V’
b’ done.’
b’’
b’Error: [NodeId: Back-Geocoding] org.jblas.NativeBlas.dgemm(CCIIID[DII[DIID[DII)V’
b’-- org.jblas INFO Deleting /tmp/jblas2130262475707773817/libjblas.so’
b’-- org.jblas INFO Deleting /tmp/jblas2130262475707773817/libgfortran-4.so’
b’-- org.jblas INFO Deleting /tmp/jblas2130262475707773817/libjblas_arch_flavor.so’
b’-- org.jblas INFO Deleting /tmp/jblas2130262475707773817/libquadmath-0.so’
b’-- org.jblas INFO Deleting /tmp/jblas2130262475707773817’
033[1;35m Error in processing … coreg_ifg2run
I don’t fix my probem, I need help
Thanks.
As I have not developed or tested this script, I cannot help much.
But there is a wiki article on this type of error: https://senbox.atlassian.net/wiki/spaces/SNAP/pages/321126402/Collection+of+FAQs#On-Linux-I-get-the-error-org.jblas.NativeBlas.dgemm(CCIIID[DII[DIID[DII)V
I met the same error, and I tried to install libgfortran.so.5, but another error occured and I’m now still stuggling with it.
Hi @dongyusen
I loaded the ‘sbas_topsar_coreg.xml’ graph and found that there was no operator connected after ‘enhance spectral diversity’
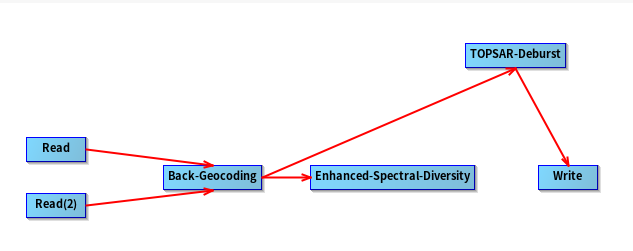
Could you please explain why the graph is designed like that? (I’m a beginner in InSAR, maybe I asked a stupid question  )
)
Thank you in advance
I have processed 56 Sentinel 1 data with sbas_snap2stamps and it worked fine, but there are some things to note:
This is different from snap2stamps which separate slave and master SLC data folders, sbas_snap2stamp combines master and slave file folders.
in the configure file, the AOI clip area must be filled in properly, the size must not be more than AOI BBOX DEFINITION.
The order of work:
check the baseline with plotbaseline.py, if something is not connected, you can add it to the sbas_add file, then run sbas_topsar_ifg by taking longer temp_baseline on the configure file.
When setting mt_prep_snap rg_patch and az_ptch, for example:
mt_prep_snap 20150119 / min / g / process / stamps / bandung_basin / 149_615 / SBAS / INSAR_20150119 / SMALL_BASELINES / 20141108_20151103 / 0.6 5 5 50 200
this is because all data can be stored and processed.
This is most important, if the stamps are downloaded from https://github.com/dbekaert/StaMPS/ when the sb_parm_initial command is activated or small_baseline_flag: ‘y’ in stamps (1), stamps step 1 will occur an error.
download and replace the sb_load_initial_gamma.m file from https://github.com/limingjia92/StaMPS/blob/patch-2/matlab/sb_load_initial_gamma.m from the matlab folder.
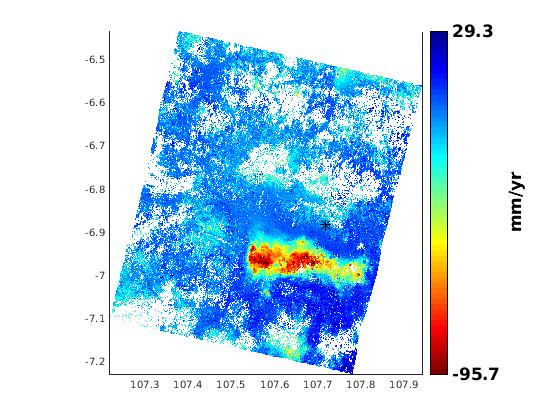
sbas result
.
.
.
and I already process with ps insar
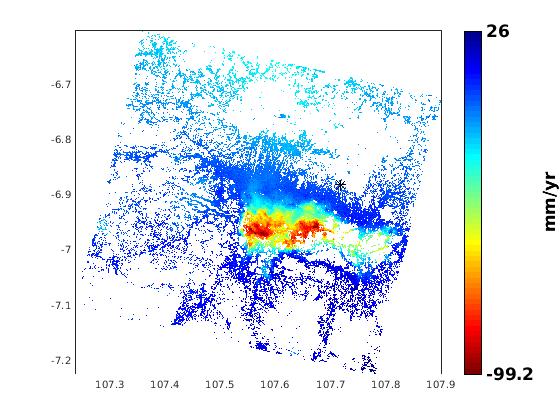
ps result
I had update the sbas_topsar_coreg.xml file, please download the new file.
thx!