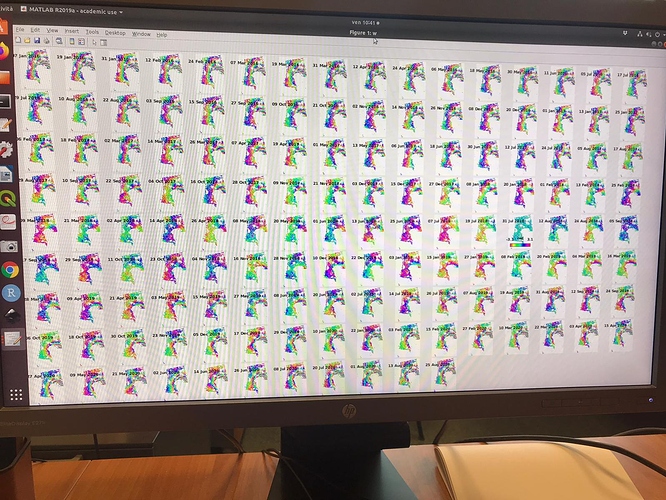
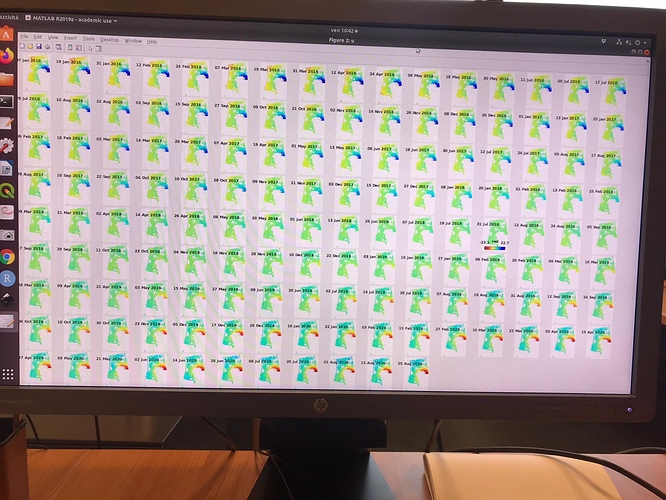
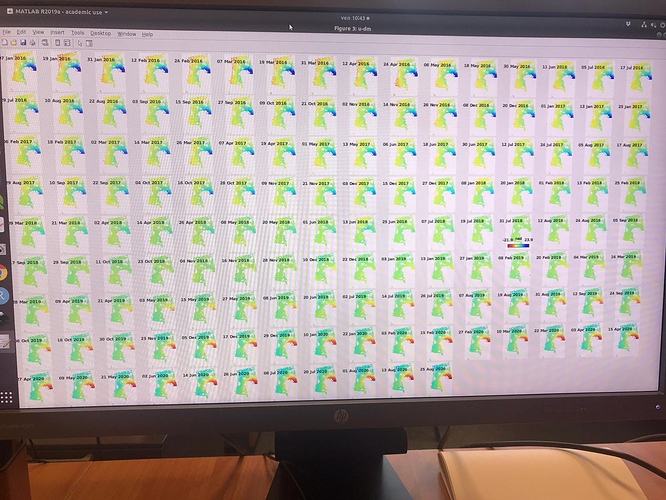
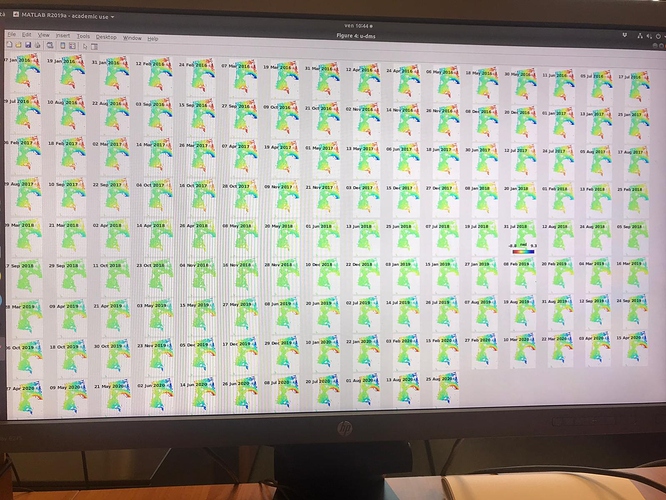
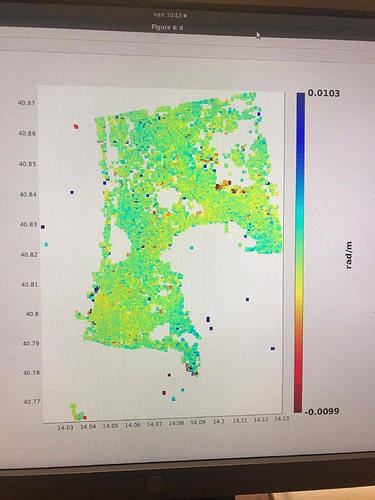
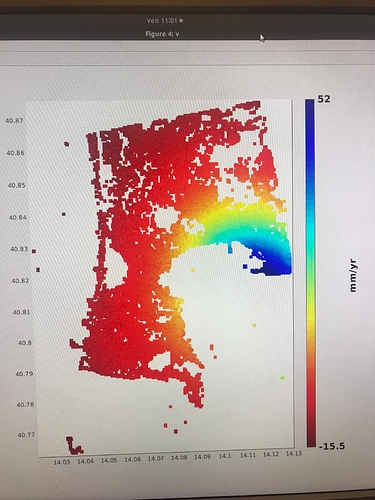
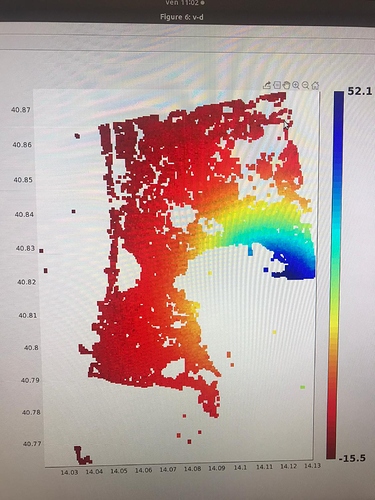
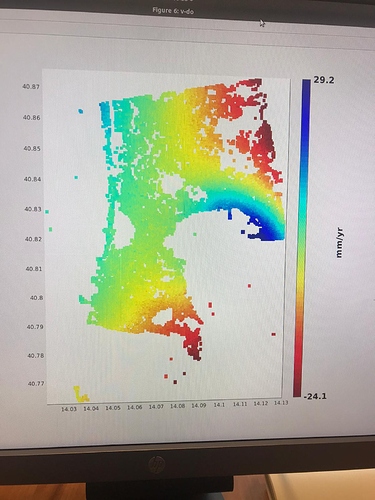
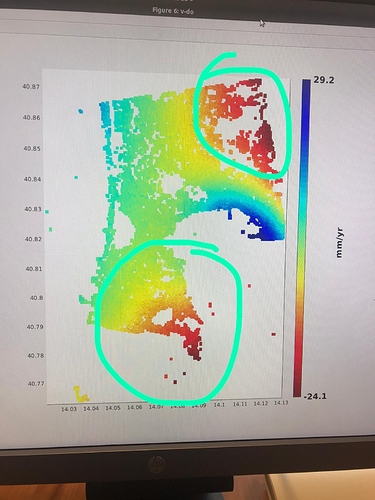
I did run it and I found no issues on that AOI. Which issue was given in step7?
Hello
in the elaboration I noticed results that are different from those expected in the various articles. Especially in the areas circled in green in the last image. it seems to eliminate some of the deformation.
I have attached the plots.
Do you have any suggestions?
many Thanks
Francesco Menniti
Hi
I run the python sbas_topsar_ifg.py project.txt command but it seems to have not created the IFGs.
I continued with the python sbas_export.py project.txt command and the command seems to work fine.
When I run the command mt_prep_snap … it gives me this mean amplitude 0.
pening /home/protepos/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES/20180108_20200813/rslc/20180108.rslc…
WARNING : SLC /home/protepos/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES/20180108_20200813/rslc/20180108.rslchas ZERO mean amplitude
Mean amplitude = 0
And when I run stamps(1,5) i get this error
Index in position 2 exceeds array bounds.
Error in sb_load_initial_gamma (line 97)
rg=rgn+ij(:,3)*rps;
Error in stamps (line 222)
sb_load_initial_gamma;
can you help me @dongyusen @ABraun
thank you very much for your availability
please list the exact mt_prep_snap command you used
I have used this comand
/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES/20180108_20200813$ mt_prep_snap 20180508 /home/protepos/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES/20180108_20200813 0.65 1 3
I think you should run it in the SMALL_BASELINES directory, not in one of the image pairs’ folder:
![]()
even with this command the result does not change!
it seems he didn’t create the interferograms
protepos@protepos:~/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES$ mt_prep_snap 20180508 /home/protepos/ELABORAZIONE_BAIA_SNAP_SBAS/ASC/INSAR_20180508/SMALL_BASELINES/20180108_20200813 0.65 3 2
did you visually check some of the interferograms
yes, check one if they contain valid data
I have checked the .png and all .png are empty.
I have no idea what went wrong then - we will have to wait for @dongyusen who delveloped the package.
your working looks fine, but the result is unacceptable. Maybe it’s cauced by the atmospheric errors.
please check interferograms with snap. there are two reasons for the blank .png:
(1) check your python: import matplotlib.pyplot as plt
(2) errors in data processing.
By the way, do you download the newest version from github ?
Thanks for reply!
what exactly should I have to modify into line 141 in sb_load_initial_gamma.m?
Yes I have download a new version from github!
Monday I will check the ifs!
Thank you very much
@volter29 How about your test ?
That’s right!
sorry but for business reasons I had to stop processing.
I will resume the test in a couple of weeks.