Well done! Thanks @panggea.
the scripts are working on python3.8 and snap7.0, please test the snap8.0. thx
Good afternoon,
I am using PSI and now i would like to try this new method SBAS, but i cannot understand what cropSx,CropSy…etc means. if you can give me some examples.
In my opinion, cropSx means ‘Startx’, cropSy means ‘Starty’, and CropWx(y) means the width and hight of the image after subset. I checked these from gpt Subset -h, I don’t know whether my understanding is correct. 
Hi @Linya-Peng , @dongyusen
Please we need more clarification and how to obtain them
CropSx=? , CropSy=? , CropWx=? , CropWy=?
Thanks.
I used the subset operator in SNAP GUI to obtain them
Dear @dongyusen
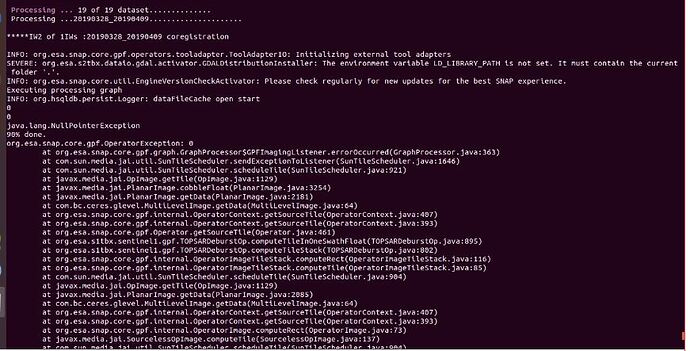
In the step sbas_topsar_coreg ,I get the error bellow and I fail to fix it
Processing … 4 of 4 dataset…
Processing …20161220_20170302…
*****IW2 of 1IWs :20161220_20170302 coregistration
b’INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters’
b"SEVERE: org.esa.s2tbx.dataio.gdal.activator.GDALDistributionInstaller: The environment variable LD_LIBRARY_PATH is not set. It must contain the current folder ‘.’."
b’INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.’
b’Executing processing graph’
b’INFO: org.hsqldb.persist.Logger: dataFileCache open start’
b"…10%…20%…Java HotSpot™ 64-Bit Server VM warning: INFO: os::commit_memory(0x00007fc9a4580000, 466092032, 0) failed; error=‘Ne peut allouer de la m\xc3\xa9moire’ (errno=12)"
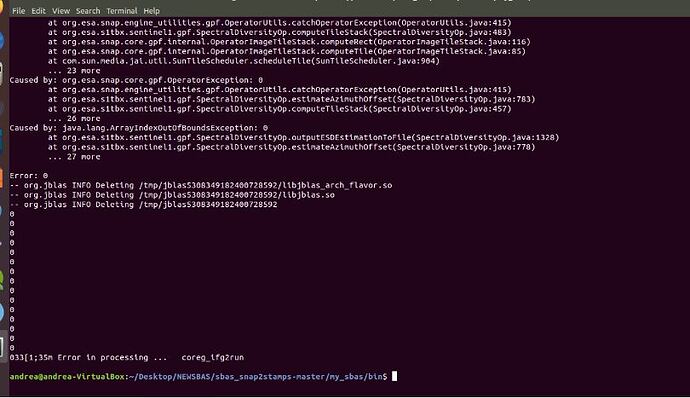
b’#’
b’# There is insufficient memory for the Java Runtime Environment to continue.’
b’# Native memory allocation (mmap) failed to map 466092032 bytes for committing reserved memory.’
b’# An error report file with more information is saved as:’
b’# study/my_sbas/bin/hs_err_pid6633.log’
033[1;35m Error in processing … coreg_ifg2run
hs_err_pid6633.log (141.9 KB)
this is quite clear I would say…
Hi @dongyusen ! Sorry to disturbe~
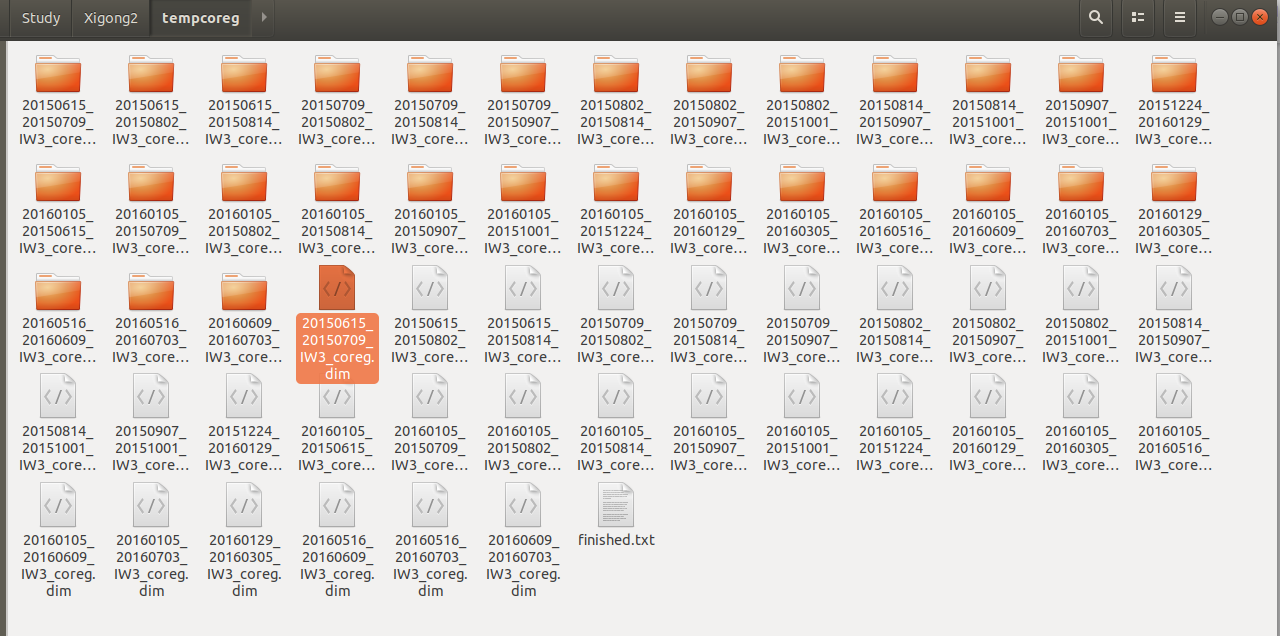
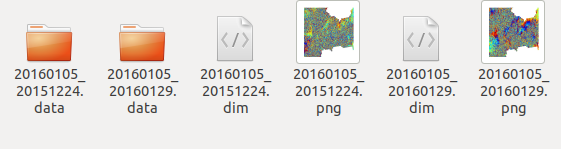
I finished the “sbas_topsar_coreg.py”, and got 29 coregistration pairs in the tempcoreg folder:
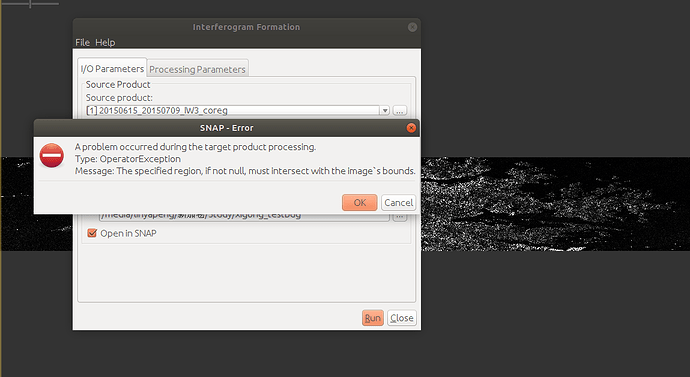
All of them are not empty, and could be opened in SNAP GUI. However, I got a “the specified region if not null, must intersect with the image’s boounds” error while running “sbas_topsar_ifg.py”. I tried to form ifg in SNAP and still got the same error:
However, some ifgs successfully generated, like this:
I search the problem in the forum but the situation is not same, could you please help me?
but I have sufficient memory , here why I didn’t understand
sorry, I don’t know your computer’s capabilities, so I was just interpreting the error message.
I have a mistake but I don’t know what it is, someone can bend the why?
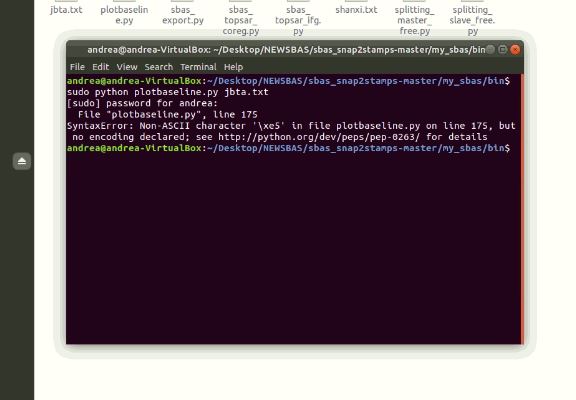
please open the script with a text editor, go to line 175 (third from the bottom) and remove the asian characters, then save the file and run the script again.
which SNAP version are you using? This should be solved since quite a time.
I’m using snap 6.0? This is the problem?
yes, please install version 8 and all updates.
Hi! @dongyusen
I think I found the solution to the error( “the specified region if not null, must intersect with the image’s boounds”) I mentioned above , and it may be something relate to the sbas_topsar_coreg.py.
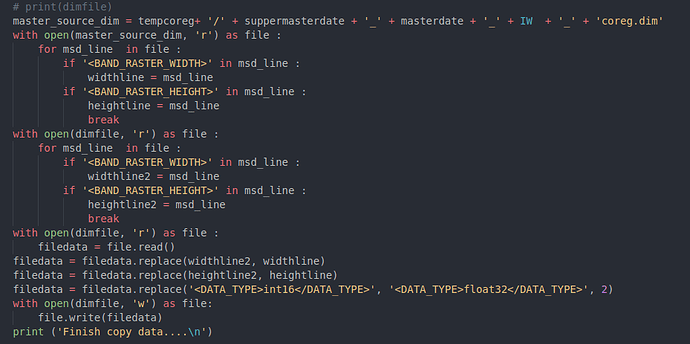
In the modify the .dim file part
The script only modifies the <BAND_RASTER_WIDTH> and <BAND_RASTER_HEIGHT>. However, in the .dim file, there is a <Raster_Dimensions> parameter may also need to be modified.

I tried to manually redo the whole .dim file modification process, same as the script, then the error took place again when I tried to form ifg in SNAP GUI.But when I modified the <Raster_Dimensions> manually, then the ifg formation tool could work.
I have no idea why this wired error took place in my work, while other SAR people’s could work fine. Do you have any idea?
Thank you again for sharing such wonderful scripts!

thank you for working on the tops_coreg script, i also get the same problem.
but I cannot understand on what basis you have modified the file dim, can you help me?
Sorry, I’m not a native speaker in English, so I didn’t get your point.  Do you mean which tool I used to edit the .dim file? or what parameter I modified?
Do you mean which tool I used to edit the .dim file? or what parameter I modified?