i using snap 8.0 and python 2.7 and i have the error mentioned above. i tried your advice but i have the same error, ihave the same coordinates in THE CLIP AOI and i change the AOI LONG E LAT MORE EXTENDED.
I don’t belive that is the problem of python version @ABraun
I have never said that ![]()
thank you for this note. How exactly should a missing pair be added to the sbas_add file? I’m not quite sure about the formatting.
Edit: Nevermind, I found it out. It should look like this
2015111920151225
2015122520160130
2016021120160318
2016031820160423
2016072820160902
Simply a combination of both dates. Once these are added, I updated the baseline in the config file and ran both scripts again, sbas_topsar_coreg and sbas_topsar_ifg. Or is this too much to manually add connections?
Script works well so far, thank you @dongyusen
No, your step is right @ABraun.
But you should pay attention to time baselines in sbas_topsar_ifg, you may have to change them to longer according to the maximum time difference of the interferogram pairs you add. If you don’t change, it could be that the interferogram pair you added will not be processed in sbas_topsar_ifg.
Hi ABraun!
May I ask which SNAP version and python version did you use to run the script? I still couldn’t form ifg after successfully finishing coregistration, only coreg pairs of supermaster and slave could form ifg successfully. I don’t know whether it is a version problem or I did something wrong 
SNAP 8.0.3 Python 3.8
the error during the interferogram computation is caused by the Land Sea Mask operator which tries to download SRTM 3Sec data as well. Updating SNAP and deleting incomplete zip files should help, otherwise you can remove the Land Sea Mask operator from the XML (sbas_topsar_1iw-seamask.xml, “Read” should directly feed into “Inteferogram”)
I removed the Land Sea Mask operator, but the error still existed. However, when I modified the NCOLS and NROWS from <Raster_Dimensions> in .dim file, and made their values same as <BAND_RASTER_WIDTH> and <BAND_RASTER_HEIGHT>, then the ifg can formed. I can’t understand why this problem took place, and whether the ifg results were right  ,Do you have any idea?
,Do you have any idea?
Thank you in advance~
are you sure? It is important to not only delete it in the XML but then use Read as input in the next step. This allowed me to proceed with this step.
About the NCOLS and NROWS - this is what worked for me:
- TOPS Split
- TOPS Deburst
- Subset to the area of interest (noting down the pixel extents [1])
- Range Doppler Terrain Correction
- Draw Rectangle around the area of interest (larger than the subset)
- WKT from Geometry (note down geographic coordinates [2])
- Enter [1] under CROP
- Enter [2] under AOI
Yes, I also used “Read” as input in next step. But for the AOI in the configure file, I just got the geographic coordinate from Google Earth, while for the Clip Area in the configure file, I just got the CropSx , CropSy, CropWx , CropWy from the Split results (Maybe this is where the error came from?)
By the way, CropSx , CropSy mean the start( Left up) pixel of the subset, and CropWx , CropWy mean the width and height of the subset, is that right?
Thank you for your patience! 
yes. I used the numbers from the subset tool, but in the end it shouldn’t matter how these were retrieved as long as there are no logical errors. Let’s wait for a fix of the StaMPS export so we can proceed.
OK~Thank you again for your patience! 
Hi ABraun! Sorry to disturbe~
I found that my Split Results didn’t share a same footprint
If 2 images are not in the same extent(for example one is bigger than another), and we didn’t subset before interferogram, will error occur?
that’s completely fine, such smaller shifts are alright.
I don’t recommend subsetting in SNAP for this task and leave it to the scripts.
CropSx , CropSy, CropWx , CropWy is necessary for sb process because the foontprint problem. when you finish sbas_topsar_coreg.py, you can load any coregisted dim from tempcoreg and find you real AOI, CropSx and CropSy are the AOI starting points in SNAP Desktop.
I see, thank you. Can the pixel and geographic coordinates rerfer to the same extent? Above, it was mentioned that the pixel extent must not be larger than the AOI extent given in coordinates. So I always made the latter slightly larger to avoid errors resulting from decimal places.
To make it short: Can we simply open a coregistered product and read both the pixel and coordinate extents from the Subset operator? Without applying it ourselves, of course.
geographic coordinates is used to clip the SAR data in splitting steps, which leads to a full swath datasets. CropSx , CropSy, CropWx , CropWy are used subset the splited dataset and make all the data with the same area.
thank you.
! New update of SBAS_snap2stamps
- bug fix,
- test for SNAP8.0 and python 3.7.9,
- add PSInSAR,
- … etc.
SNAP-SBAS-PSInSAR-StaMPS-20210206.zip (43.3 KB)
sorry, I must be stupid, but my area of interest is always shifted by several kilometers, I must be doing something wrong with the CropSx entries.
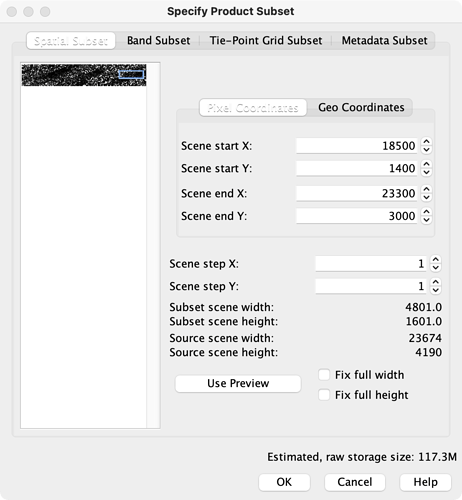
I opened one coregistered file from the folder tempcoreg [1] and zoomed to an area, then I selected Spatial Subset from View to get the pixel coordinates in the Spatial Subset tab [2]. I entered them in the config file and proceeded with the script 4sbas_topsar_ifg.py
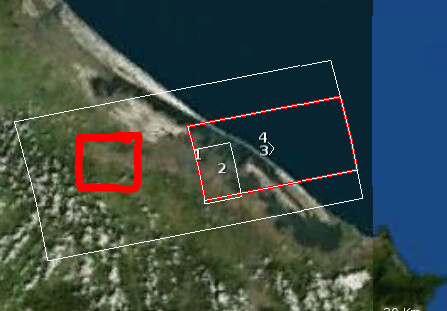
The results contain the study area [2], but the subset of the created interferograms [3 and 4] is much larger than defined in the config file
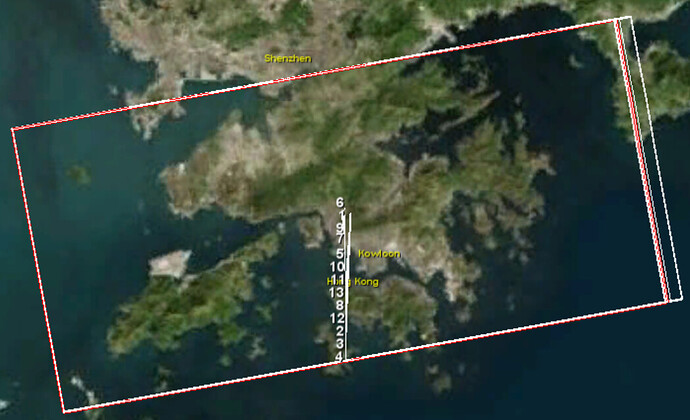
When I export to StaMPS and process the data, the results cover the red square shown below.
What am I doing wrong here?
Here is my Crop value
#AOI clip area, for data clip, the reference image should be deburst and merged, and read the x,y from co-registed file.
#The value can read from any dim in tempcoreg folder, and used to clip the coregisted files for PSInSAR/SBAS processing.
CropSx=18500
CropSy=1400
CropWx=4800
CropWy=1600
the CropSx & y read from down-right area (when mouse point on the start point),
be care the start end in “Specify Product Subset”, the CropWx = “Scene end X” - “Scence stat x”