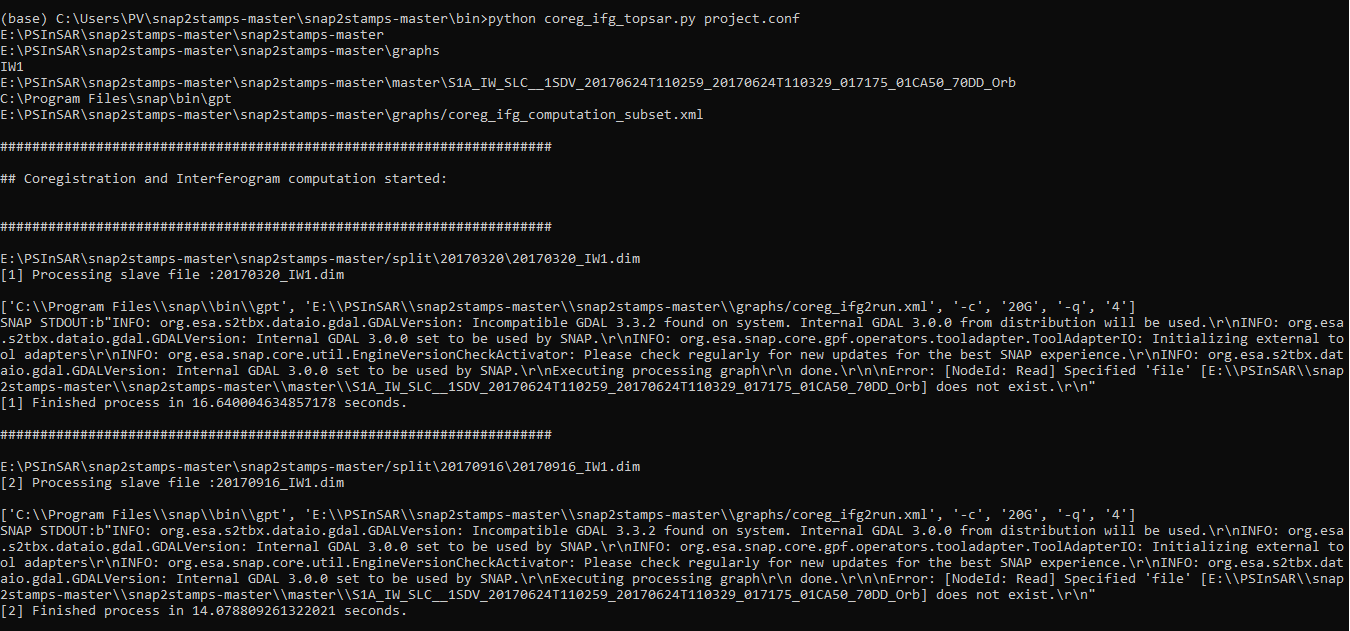
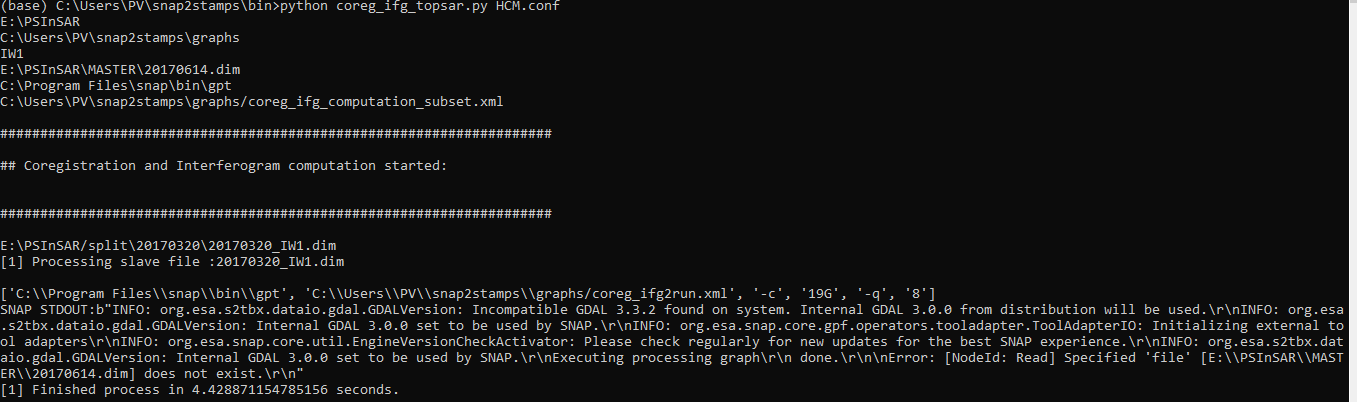
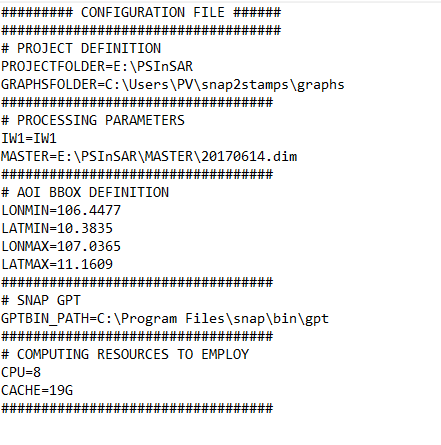
Hi, I have problem when running Coregistration and Interferogram. Please help me to solve the problem. Thank you to all
INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters\r\nINFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Incompatible GDAL 3.3.2 found on system. Internal GDAL 3.0.0 from distribution will be used.\r\nINFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.\r\nINFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.\r\nINFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.\r\nExecuting processing graph\r\n done.\r\n\nError: [NodeId: Read] Specified ‘file’
Do you use a graph to process the data? At least the error message indicates it.
If so, please share the XML of the graph in here and describe how you run the graph (e.g. from inside SNAP or via gpt)
I use snap2temps for data preparation. I run by python 3.8.10 in terminal of anaconda (the process via gpt ). Although before, I aslo pass though the step for Coregistration and Interferogram, when i run again, it doesn’t work.
Is E: and external storage (e.g., USB or network drive)? Do you have long paths enabled? Not all external storage supports long paths.