Hi again everyone,
I am trying to make a mosaic from S1A products. I managed to go a bit further with the whole procedure but now I am getting some strange stuff while mosaicing. I will start with the products and the code:
import sys
import snappy
from snappy import ProductIO
from snappy import HashMap
import os
from snappy import GPF
from pyproj import Proj
import osgeo.osr
import osr
import gdal, gdalconst
import glob
GPF.getDefaultInstance().getOperatorSpiRegistry().loadOperatorSpis()
HashMap = snappy.jpy.get_type('java.util.HashMap')
safe_files = [S1A_EW_GRDM_1SDH_20170222T082610_20170222T082710_015394_01942C_36A4.SAFE',
S1A_EW_GRDM_1SDH_20170222T082510_20170222T082610_015394_01942C_A081.SAFE',
S1A_EW_GRDM_1SDH_20170222T082410_20170222T082510_015394_01942C_945D.SAFE']
proj4str = "+proj={0} +lat_0={1} +lon_0={2} " \
"+lat_ts={3} +ellps={4} +datum={5} +units={6} " \
"+lon_wrap={7}".format("stere",85,47,85,"WGS84","WGS84","m",0)
srsproj4 = Proj(proj4str)
srs = osgeo.osr.SpatialReference()
srs.ImportFromProj4(srsproj4.srs)
def do_calibration(source, polarization):
"""
radiometric calibration
"""
parameters = HashMap()
parameters.put('outputSigmaBand', True)
parameters.put('sourceBands', 'Intensity_' + polarization)
parameters.put('selectedPolarisations', polarization)
parameters.put('outputImageScaleInDb', False)
target_0 = GPF.createProduct("Calibration", parameters, source)
return target_0
def do_speckle_filtering(source):
"""
"""
parameters = HashMap()
parameters.put('sourceBandNames', 'Sigma0_HH')
parameters.put('filter', 'Refined Lee')
output = GPF.createProduct('Speckle-Filter', parameters, source)
return output
def do_terrain_correction(source, crs):
"""
"""
outname = source.getDisplayName()
outname = outname + '_tc'
parameters = HashMap()
parameters.put('demName', 'GETASSE30') #ASTER 1Sec GDEM
parameters.put('demResamplingMethod', 'BILINEAR_INTERPOLATION')
parameters.put('imgResamplingMethod', 'BILINEAR_INTERPOLATION')
parameters.put('pixelSpacingInMeter', 150.0)
parameters.put('sourceBands', 'Sigma0_HH')
parameters.put('incidenceAngleForSigma0', 'Use incidence angle from Ellipsoid')
parameters.put('mapProjection', crs)
parameters.put('nodataValueAtSea', False)
output = GPF.createProduct('Terrain-Correction', parameters, source)
return output
def remove_thermal_noise(source, polarization):
"""
"""
parameters = HashMap()
parameters.put('selectedPolarisations', polarization)
output = GPF.createProduct('ThermalNoiseRemoval', parameters, source)
return output
def remove_GRD_border_noise(source, polarization):
"""
removes the black areas around the image
Mask no-value pixels for GRD product
"""
parameters = HashMap()
parameters.put('selectedPolarisations', polarization)
output = GPF.createProduct('Remove-GRD-Border-Noise', parameters, source)
return output
def do_mosaic(source):
"""
"""
parameters = HashMap()
parameters.put('pixelSize', 150.0)
parameters.put('resamplingMethod', 'BILINEAR_INTERPOLATION')
output = GPF.createProduct('SAR-Mosaic', parameters, source)
return output
# main script
for s1file in safe_files:
print(s1file)
sentinel_1 = ProductIO.readProduct(s1file + "/manifest.safe")
polarization = 'HH'
TNfiltered = remove_thermal_noise(sentinel_1, polarization)
BNfiltered = remove_GRD_border_noise(TNfiltered, polarization)
calibrated = do_calibration(BNfiltered, polarization)
filtered = do_speckle_filtering(calibrated)
tercorrected = do_terrain_correction(filtered, srs.ExportToWkt())
ProductIO.writeProduct(tercorrected, sentinel_1.getName()+'_final', 'GeoTIFF')
sentinel_1.dispose()
sentinel_1.closeIO()
mosaic_files = glob.glob(os.path.join('/home/pakoun/workspace/image_correction', '*final*'))
products = []
for fl in mosaic_files:
products.append(ProductIO.readProduct(s1file + "/manifest.safe"))
mosaic = do_mosaic(products)
ProductIO.writeProduct(mosaic, 'mosaic', 'GeoTIFF')
When running the Mosaic i get the following warning:
first_line_time metadata value is null
last_line_time metadata value is null
Nevertheless it seems that my file is created. Now if I load it in qgis the mosaic is the (somehow) colored one. As you see this is not a mosaic at all but just only 1 acquisition. Although it supposed to be also TC it looks like it is not projected. For comparison I am plotting also another acquisition (black and white picture) which I performed exactly the same analysis procedure but no Mosaicing. Interestingly enough although I performed a GRD_border_noise removal, the black background which corresponds for the NAN values is present. Further there is a misalignment with the shore which is around 6km…
There might be lot of questions in only one post but I though it would be of great help (providing also the code) for others that they are interested to follow the same procedure.
I trully hope for your input cause I am struggling to understand what is wrong here…

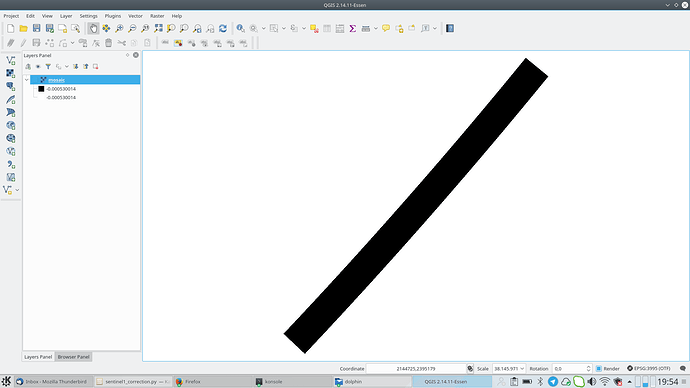
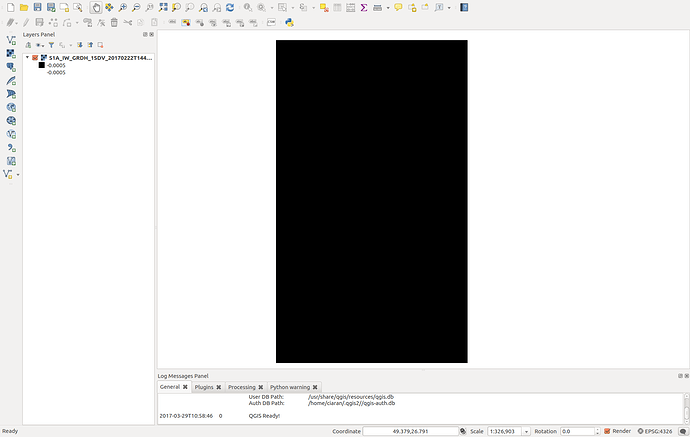
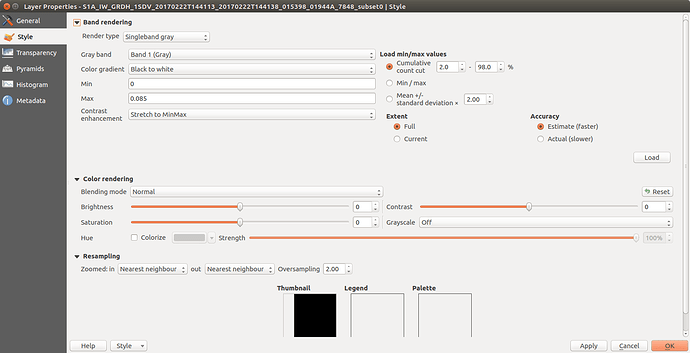
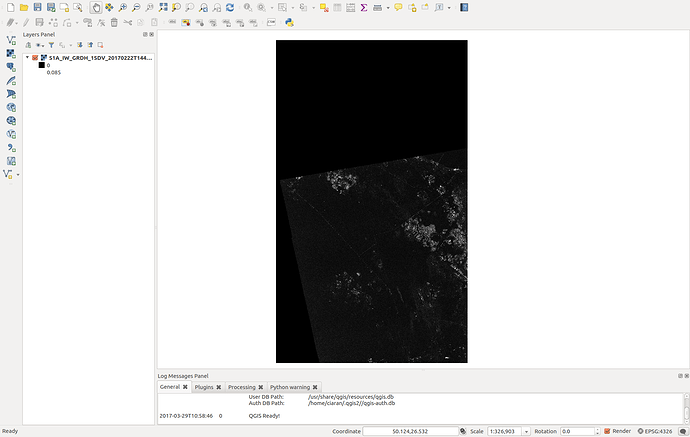
 ) The second weird thing is that from this noise that I retrieved, was showing only 1 out of the 3 products I used. Nevertheless you see here a very wide result. So basically that was not a mosaic. I am just thinking if this is caused due to NAN values around the acquisition (thats the black area when you insert ESA S1 product directly to qgis). If so, somehow those should be depricated; something similar to the gdal command :
) The second weird thing is that from this noise that I retrieved, was showing only 1 out of the 3 products I used. Nevertheless you see here a very wide result. So basically that was not a mosaic. I am just thinking if this is caused due to NAN values around the acquisition (thats the black area when you insert ESA S1 product directly to qgis). If so, somehow those should be depricated; something similar to the gdal command :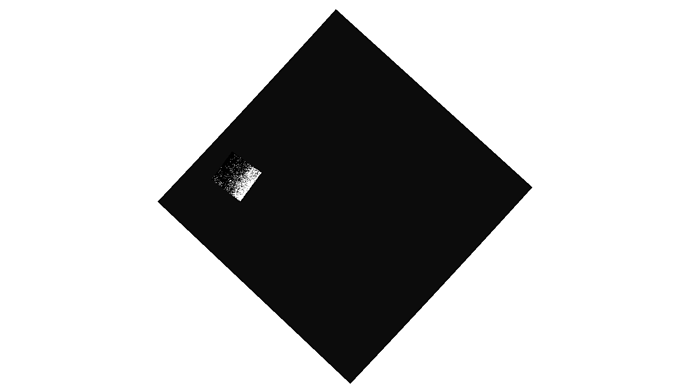
 Its definitely coarser but still the spatial structure I want to see is resolved. Consider also that if you want to plot a jpeg or something, even if you have 10m resolution, the dpi that you define when saving a plot most likely, will not be able to resolve the nominal spatial resolution.
Its definitely coarser but still the spatial structure I want to see is resolved. Consider also that if you want to plot a jpeg or something, even if you have 10m resolution, the dpi that you define when saving a plot most likely, will not be able to resolve the nominal spatial resolution.