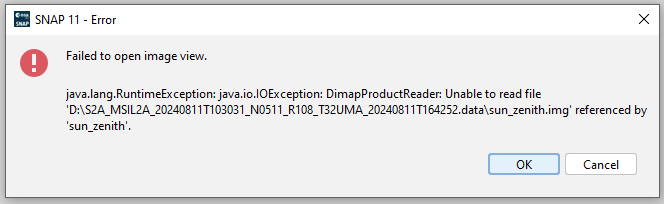
Is it possible to read all bands, metadata, and ancillary data of Sentinel-2 by reading its manifest.safe or MTD_MSIL2A.xml?
Example:
- For reading
MTD_MSIL2A.xmlfile
import esa_snappy
from esa_snappy import ProductIO
data = ProductIO.readProduct('D:/S2A_MSIL2A_20240811T103031_N0511_R108_T32UMA_20240811T164252.SAFE/MTD_MSIL2A.xml')
RuntimeError: java.lang.NoClassDefFoundError: Could not initialize class org.gdal.gdalconst.gdalconstConstants
- For reading
manifest.safefile, its read without any error but as an empty object
import esa_snappy
from esa_snappy import ProductIO
data = ProductIO.readProduct('D:/S2A_MSIL2A_20240811T103031_N0511_R108_T32UMA_20240811T164252.SAFE/manifest.safe')
bands = list(data.getBandNames())
bands
AttributeError: 'NoneType' object has no attribute 'getBandNames'
- After converting the product into
BEAM-DIMAPformat, e.g.,
import os
import esa_snappy
from esa_snappy import ProductIO
product = ProductIO.readProduct("D:/S2A_MSIL2A_20240811T103031_N0511_R108_T32UMA_20240811T164252.SAFE")
output = ProductIO.writeProduct(product, "D:/S2A_MSIL2A_20240811T103031_N0511_R108_T32UMA_20240811T164252.dim", "BEAM-DIMAP")
esa_snappy module can list all data
data = ProductIO.readProduct('D:/S2A_MSIL2A_20240811T103031_N0511_R108_T32UMA_20240811T164252.dim')
bands = list(dataSEN2L2.getBandNames())
bands
[Output:
'B1',
'B2',
'B3',
'B4',
'B5',
'B6',
'B7',
'B8',
'B8A',
'B9',
'B11',
'B12',
'quality_aot',
'quality_wvp',
'quality_cloud_confidence',
'quality_snow_confidence',
'quality_scene_classification',
'B_detector_footprint_B1',
'B_detector_footprint_B2',
'B_detector_footprint_B3',
'B_detector_footprint_B4',
'B_detector_footprint_B5',
'B_detector_footprint_B6',
'B_detector_footprint_B7',
'B_detector_footprint_B8',
'B_detector_footprint_B8A',
'B_detector_footprint_B9',
'B_detector_footprint_B11',
'B_detector_footprint_B12',
'B_ancillary_lost_B1',
'B_ancillary_degraded_B1',
'B_msi_lost_B1',
'B_msi_degraded_B1',
'B_defective_B1',
'B_nodata_B1',
'B_partially_corrected_crosstalk_B1',
'B_saturated_l1a_B1',
'B_ancillary_lost_B2',
'B_ancillary_degraded_B2',
'B_msi_lost_B2',
'B_msi_degraded_B2',
'B_defective_B2',
'B_nodata_B2',
'B_partially_corrected_crosstalk_B2',
'B_saturated_l1a_B2',
'B_ancillary_lost_B3',
'B_ancillary_degraded_B3',
'B_msi_lost_B3',
'B_msi_degraded_B3',
'B_defective_B3',
'B_nodata_B3',
'B_partially_corrected_crosstalk_B3',
'B_saturated_l1a_B3',
'B_ancillary_lost_B4',
'B_ancillary_degraded_B4',
'B_msi_lost_B4',
'B_msi_degraded_B4',
'B_defective_B4',
'B_nodata_B4',
'B_partially_corrected_crosstalk_B4',
'B_saturated_l1a_B4',
'B_ancillary_lost_B5',
'B_ancillary_degraded_B5',
'B_msi_lost_B5',
'B_msi_degraded_B5',
'B_defective_B5',
'B_nodata_B5',
'B_partially_corrected_crosstalk_B5',
'B_saturated_l1a_B5',
'B_ancillary_lost_B6',
'B_ancillary_degraded_B6',
'B_msi_lost_B6',
'B_msi_degraded_B6',
'B_defective_B6',
'B_nodata_B6',
'B_partially_corrected_crosstalk_B6',
'B_saturated_l1a_B6',
'B_ancillary_lost_B7',
'B_ancillary_degraded_B7',
'B_msi_lost_B7',
'B_msi_degraded_B7',
'B_defective_B7',
'B_nodata_B7',
'B_partially_corrected_crosstalk_B7',
'B_saturated_l1a_B7',
'B_ancillary_lost_B8',
'B_ancillary_degraded_B8',
'B_msi_lost_B8',
'B_msi_degraded_B8',
'B_defective_B8',
'B_nodata_B8',
'B_partially_corrected_crosstalk_B8',
'B_saturated_l1a_B8',
'B_ancillary_lost_B8A',
'B_ancillary_degraded_B8A',
'B_msi_lost_B8A',
'B_msi_degraded_B8A',
'B_defective_B8A',
'B_nodata_B8A',
'B_partially_corrected_crosstalk_B8A',
'B_saturated_l1a_B8A',
'B_ancillary_lost_B9',
'B_ancillary_degraded_B9',
'B_msi_lost_B9',
'B_msi_degraded_B9',
'B_defective_B9',
'B_nodata_B9',
'B_partially_corrected_crosstalk_B9',
'B_saturated_l1a_B9',
'B_ancillary_lost_B11',
'B_ancillary_degraded_B11',
'B_msi_lost_B11',
'B_msi_degraded_B11',
'B_defective_B11',
'B_nodata_B11',
'B_partially_corrected_crosstalk_B11',
'B_saturated_l1a_B11',
'B_ancillary_lost_B12',
'B_ancillary_degraded_B12',
'B_msi_lost_B12',
'B_msi_degraded_B12',
'B_defective_B12',
'B_nodata_B12',
'B_partially_corrected_crosstalk_B12',
'B_saturated_l1a_B12',
'B_opaque_clouds',
'B_cirrus_clouds',
'B_snow_and_ice_areas',
'view_zenith_mean',
'view_azimuth_mean',
'sun_zenith',
'sun_azimuth',
'view_zenith_B1',
'view_azimuth_B1',
'view_zenith_B2',
'view_azimuth_B2',
'view_zenith_B3',
'view_azimuth_B3',
'view_zenith_B4',
'view_azimuth_B4',
'view_zenith_B5',
'view_azimuth_B5',
'view_zenith_B6',
'view_azimuth_B6',
'view_zenith_B7',
'view_azimuth_B7',
'view_zenith_B8',
'view_azimuth_B8',
'view_zenith_B8A',
'view_azimuth_B8A',
'view_zenith_B9',
'view_azimuth_B9',
'view_zenith_B10',
'view_azimuth_B10',
'view_zenith_B11',
'view_azimuth_B11',
'view_zenith_B12',
'view_azimuth_B12']