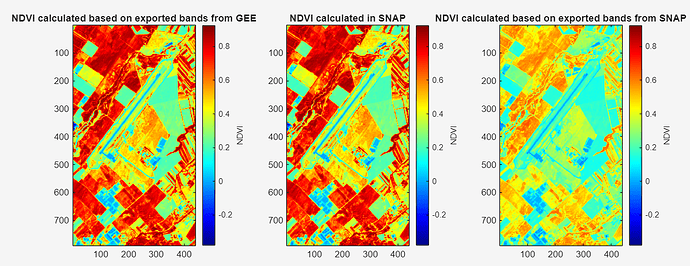
When I use SNAP’s NDVI processor and Band Math I obtain different values than the calculation in Matlab (or Python) after exporting the necessary bands. SNAP applies corrections during the calculation and I’d like to know what those corrections are. I assume NDVI values obtained in SNAP are the correct ones, then it doesn’t give the user an option to use any other tool or programming language for robust analysis. I couldn’t find documentation that clearly describes the algorithm behind it.
after a long time issue still persists and I call it issue because if certain corrections are applied to bands during NDVI calculation in SNAP, then why those corrections are not applied to the bands when we export them? For example, if I process (resample and crop) a Sentinel-2 level-2a product in SNAP, and only select 12 main bands at subset step (cropping) for exporting by ignoring quality and other bands (I don’t have much info on them.) and then I decide to do some band math with them I would obtain wrong values for indices, usually, underestimated. I assume those additional bands I ignore at subset are the ones need to be used to correct each band but I don’t know the algorithm to do it in other programming environment. So this doesn’t give me a chance other than using SNAP for index calculation or any other band math. To strengthen my argument, I recently used Google Earth Engine’s S2 harmonized collection, and I exported .tif image that is cropped and only contains main 12 bands. Then I did NDVI calculation using red and NIR in Python and Matlab, and I obtained CORRECT NDVI values, not the underestimated ones. This means when I export bands from Google’s collection I get the corrected ones but when I export those bands using SNAP I get not corrected (or whatever the process is called) ones, thus, any calculation I do using those bands will be WRONG. And I wonder how many people already used or using this way to conduct research. I hope I’m the one who is missing a crucial step here not the software. Or maybe it is a feature, but I would like to know how to deal with it. I can prove my point by sharing a script, or visual comparison or statistical comparison or all of them.
Hello,
Your observation is quite worrying if it is true.
I might have a plausible reason for the difference between GEE and SNAP.
In SNAP the scaled geo-physical values are used and not the raw digital numbers. As far as I know you need to scale the values yourself in GEE. Please correct me if I’m wrong.
If the raw values are used, the histogram of the NDVI changes.
You can compare the raw NDVI from SNAP with the result with GEE. Maybe there are more in line.
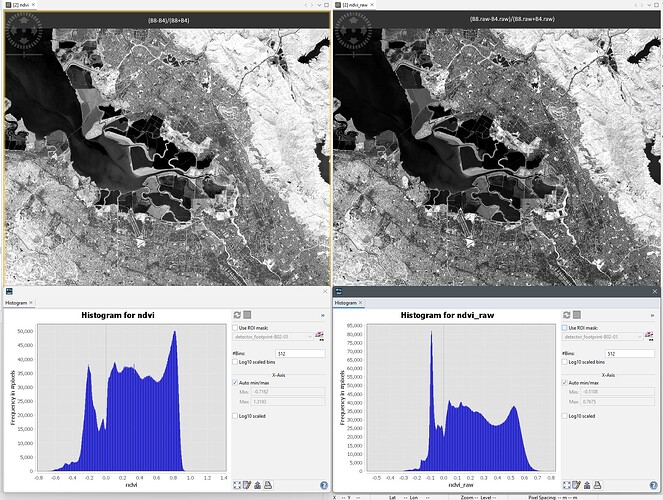
To compute the raw NDVI I’ve used the following expression:
(B8.raw-B4.raw)/(B8.raw+B4.raw)
Thanks for the reply.
Maybe images below can help to understand better.
whatever the scaling is, seems like it is applied already to the bands exported from Google Earth Engine, same goes to calculating the index inside SNAP ESA using either bandmath or already defined NDVI processor.
However, seems that it is not applied to the bands when we export from SNAP, thus, our further calculation of index is wrong.
As I understood from your reply, GEE values are not scaled, but seems like they are scaled. Not scaled or not corrected bands are the ones that the SNAP software exports.
Thanks for the details.
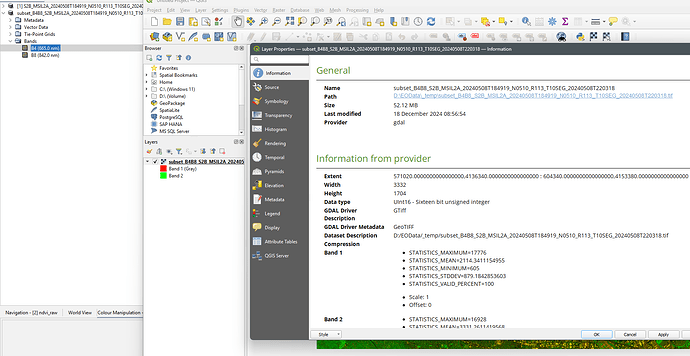
You export B4 and B8 to GeoTiff and then calculate NDVI in Python/Matlab.
So, the issue is with the export to GeoTiff and not the calculation of NDVI in SNAP. And this issue is more than 15 years old.
When exporting to GeoTiff only the raw values are exported.
You can see it in the screenshot. QGIS show UINT16 as data type.
This was a wrong decision event though there where good reasons to do so at this time.
You have 3 options, I think.
- Not using GeoTiff use NetCDF4-CF instead
- Apply the scaling and create new band in SNAP
Simply create a new band (not virtual) using BandMaths by using only the original band name. - Apply the scaling in Python/Matlab
The equation is:v = raw * factor + offset
factor = 0.0001
offset = -0.1
Thanks for the answer, I applied factor and offset and it worked. I did not say the issue is NDVI calculation in SNAP, I said issue is the calculation of NDVI in other programming environments based on the bands exported from SNAP. So I was correct on my argument that export process does something different or wrong. Maybe this makes sense considering some decisions made 15 years ago when developing the software. I saw myself how research team relied on those values until someone accidentally compared the NDVI bands. End of the day, we are users and we don’t have to be aware of every of processes that software does under the hood. Anyways, thank you very much for the solution.