Hi,
I need to install snap or s1tbx in a docker container. I use this container to download the sentinel-1 satellite images and I would to do as next step the process to regrid the satellite image.
How can I do that? It is possible to install this software on docker container? If yes, what are the instructions to do that?
Thanks in advance.
Yes, this is possible.
I think this can help you or you can use it directly: mundialis/esa-snap: Docker image of ESA Sentinel Application Platform (SNAP) (github.com)
 Marco from EOMasters - Mastering Earth Observation
Marco from EOMasters - Mastering Earth Observation
1 Like
Hi @Marco_EOM ,
I try to do it but I obtain an error. The container is not working. I follow the instructions on the Github page but is not clear how to work using Python to convert in my case images from L2 to L3. There are no information on how to do it.I would like to do that process using Python within my docker container.
Thanks in advance.
Hi @Marco_EOM,
I have finally managed to install SNAP version 9.0.0 inside my container. What is not clear to me is how to configure snappy or what are the commands from the prompt to execute. I am developing an API that is able to download images and process them making format changes (NetCDF to TIFF) to publish them in a GeoServer. The downloaded images are not well projected. My goal is something similar to what I have managed to do in the case of Sentinel-5P images. These images are L2 type and using HARP I reprojected them to be L3 type and so they are projected correctly on the map. In this case it would be the same with Sentinel1, 2 and 3 images. But the sentinel-toolbox to use is SNAP. But, I don’t know which are the commands to make this conversion. Or what is the process to do to get the same.
Thanks in advance
Hey @jclemente
great that you have managed to get the container running.
Have you already setup snappy?
If not check this wiki page: Using SNAP in your Python programs - SNAP - Confluence (atlassian.net)
If you want to do terrain correction you can use gpt.
The general API call is:
result = GPF.createProduct(operator_name, parameters, product)
By using this you can setup a chain of operations.
Depending on you needs it might be better to create a new process and call the gpt tool with a graph file instead of using the API directly. The processing can be faster.
You find some code examples on GitHub: esa-snappy/src/main/resources/esa_snappy/examples at master · senbox-org/esa-snappy (github.com)
In the Python category here in the forum are also some interesting code snipptes:
Latest development/python topics - STEP Forum (esa.int)
Hi @Marco_EOM ,
thanks for your comment and links. But I have not been able to install snappy in my container. The links refer to a traditional installation of both tools. In my case I have a container with SNAP version 9 on which I am trying to install snappy. I’ve been looking through the examples in the topic but I haven’t found any that help me with what I need. Sorry for insisting, it’s probably due to a basic problem on my part but I don’t understand the syntax and it’s not clear which tool to apply in my case. What I want is to reproject the original NetCDF and then from other packages I want to generate the TIFF of the desired parameter.
Thanks id advance
I know that installing snappy can be tricky but it general it should work in a docker container the same was other systems and explained on the wiki page.
I found another docker image. maybe this helps you out.
MATRIX4284/esa-snappy-docker: Docker for ESA SNAP Toolbox for Satellite Remote Sensing (github.com)
In general, your code should look like:
from snappy import GPF
from snappy import HashMap
from snappy import ProductIO
from snappy import jpy
inputPath = 'path/to/product'
inputProduct = ProductIO.readProduct(inputPath)
parameters = HashMap()
parameters.put('crs', 'EPSG::4326') # or other parameters
reprojected = GPF.createProduct('Reproject', parameters, inputProduct)
outputPath = 'path/to/output'
ProductIO.writeProduct(reprojected, outputPath, 'GeoTIFF-BigTIFF')
For available parameters forthe reprojection operation you can check the gpt tool on the commandline: gpt Reproject -h
Or you check the Reprojection entry in this list:
https://step.esa.int/main/wp-content/help/?version=9.0.0&helpid=gpf.operatorIndex
 Marco from EOMasters - Mastering Earth Observation
Marco from EOMasters - Mastering Earth Observation
Hi @Marco_EOM ,
None of the examples worked for me.
The docker images I have not been able to generate. It gives an error already known from what I have read in the forum and in help pages. The one that occurs when doing a snap update and the script does not finish. I have applied the indicated solution and it still does not work. In fact, I have totally changed the images to play with python versions that are indicated to be compatible but nothing. I am very lost but no solution has worked for me.
Is there any other image available?
Kind regards,
José.
Sorry I haven’t really worked with docker images yet. So, I’m also at the end of my wits.
Can someone else help here?
Please any update on that.
Can someone help me with this?
Thanks in advance.
Hi @Marco_EOM ,
After several attempts and testing with several images I can now say that I have snap and snappy in my docker container.
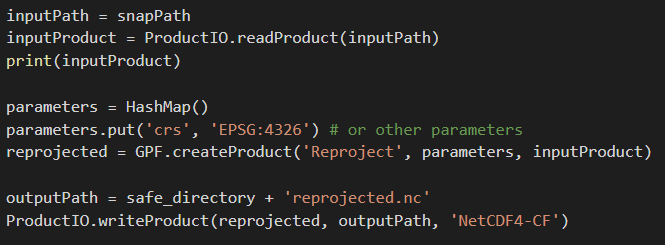
I am trying to use the code you gave me to reproject my image:
from snappy import GPF
from snappy import HashMap
from snappy import ProductIO
from snappy import jpy
inputPath = ‘path/to/product’
inputProduct = ProductIO.readProduct(inputPath)
parameters = HashMap()
parameters.put(‘crs’, ‘EPSG::4326’) # or other parameters
reprojected = GPF.createProduct(‘Reproject’, parameters, inputProduct)
outputPath = ‘path/to/output’
ProductIO.writeProduct(reprojected, outputPath, ‘GeoTIFF-BigTIFF’)
I am testing with a Sentinel-1 image (s1a-iw-ocn-vv-2024010101t055332-2024010101t055400-051909-064584-001.nc). MY goal is to reproject this NetCDF to generate another one.
I apply the previous code on this image changing only the following line:
ProductIO.writeProduct(reprojected, outputPath, ‘NetCDF4-CF’).
So that the output file becomes a NetCDF again. I don’t know if this is correct. But I get this error:
What am I doing wrong?
Thanks in advance.
Hey @jclemente great that you got it to working!
The netcdf file does not provide geo-information. The netcdf is only one component of the SAFE format. The geo-information is stored in another file.
Try to read the manifest.safe file instead, or simply the zip archive in which the nc file is located.
The NetCDF file does not contain geo-information which is usable right away. Thus, the specific reader for S1 L2 OCN data is needed.
@Marco_EOM ,
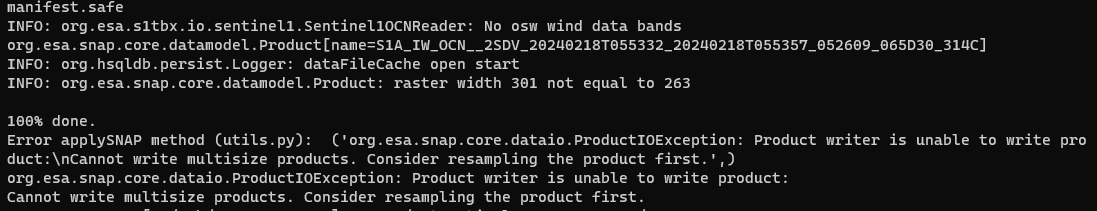
I have repeated the process but reading the manifest.safe file as you indicated.
I got the following error:
Here I think I have a concept problem. What I intended was to obtain a NetCDF that could contain all the parameters of the original NetCDF but well reprojected. Seeing the error I understand that this is not correct.
What extension should my output file have? Should I do the reprojection parameter by parameter?
I’m sorry to hear that. I don’t know the format well. But by looking at the data in SNAP Desktop most of the bands are wrong/useless. For most bands there are five variations EW1 to EW5. In SNAP they all have the same data. I doubt this is correct.
But all bands containing owi in the name seem to be good.
Maybe @jun_lu or @lveci can comment on this?
If you do a subset before the reprojection it should work.
parameters = HashMap()
bands= 'hh_001_owiRadVel,hh_001_owiLon, ...'
# or you can set this up by getting all bands names and checking for owi
bands = filterBandNames(inputProduct.getBandNames())
parameters.put(‘sourceBands’, bands)
subsetProduct = GPF.createProduct('Subset', parameters, inputProduct)
...
reprojected = GPF.createProduct('Reproject', parameters, subsetProduct)
...
def filterBandNames(bandNames):
filtered_names = [name for name in bandNames if 'owi' in name]
return ','.join(filtered_names)
Thanks a lot @Marco_EOM ,
following your recommendations I have been able to reproject the Sentinel-1 images filtering by the bands I am interested in.
I am trying to replicate the same with Sentinel-2 but when I try to read the bands from the manifest.safe file it always returns no bands.
Does the process change something when they are Sentinel-2 images?
Use the MTD_MSIL1C.xml or the MTD_MSIL2A.xml instead, depending on the level. Alternatively, you can use the zip the S2 data is distributed.
You probably need to resample the data before to make the resolution of the bands equal.
You can use the generic resampling or the S2 specific resampling. The specific one is more precise for S2 data, but you need to decide if you need this precision. It costs more computation time.
GPF.createProduct('Resample', parameters, product)
or
GPF.createProduct('S2Resampling', parameters, product)
See:
https://step.esa.int/main/wp-content/help/?version=9.0.0&helpid=s2resampler
For the operator specific parameters
you can call gpt
gpt Resample -h
or
gpt S2Resampling -h
@Marco_EOM ,
I have made the test reading the xml files that you indicate in the case of Sentinel-2. But I get the same error: ‘Object has no attribute getBandNames’.
In fact I wonder what would happen in the case of Sentinel-3. I understand that each time it is a different file that must be processed to recover the bands. Which one is for Sentinel-3?
For Sentinel-3 it is the file: xfdumanifest.xml
However, the message ‘Object has no attribute getBandNames’ indicates that getBandNames() is not invoked on a Product object.
Can you show the related code snippet?
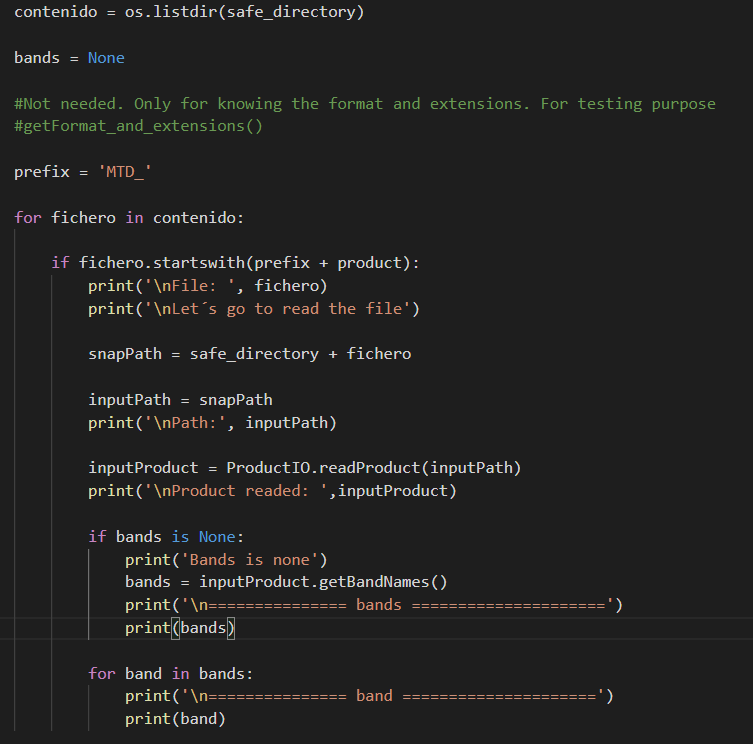
@Marco_EOM this is my code:
As you see I use MTD as prefix to read the specific file you told me for Sentinel-2. Product could be: MSIL1C, MSIL2A.
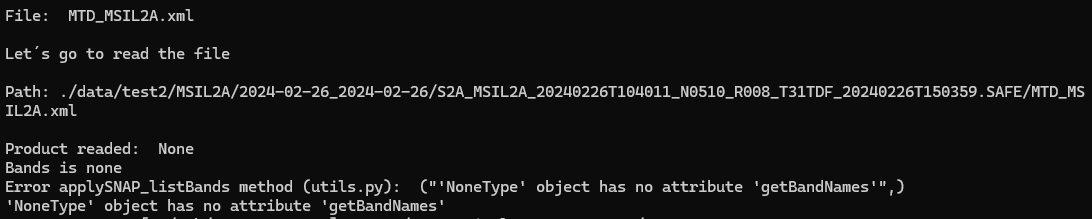
The next log shows that I read the right file but I obtain there are no information for the bands.
Variable file contains the name of the file.
Variable path is the full path including folder and file (absolute route) and as a result I obtain None.
The same for both products (MSIL1C, MSIL2A)
Something must be wrong either with the path or the S2 product.
Because it is not read. None is returned. That’s also the reason the getBandNames() method is not found.
None is returned if no reader is found for the file. Can you try to read the same product in SNAP Desktop by using the MTD_MSIL2A.xml file. Maybe the S2 Toolbox is not installed? This could be a reason for this behaviour.
And can you check the path if it is correct? Maybe by converting it to an absolute path.
import os
absPath = os.path.abspath(inputPath)
print(abspath)