When I read S2A_OPER_PRD_MSIL1C_PDMC data in s2tbx, the tool gives error messsage : java.lang.NullPointerException.
How I can fix this error message?
What version of SNAP are you using? If you don’t have the 2.0 Beta-5 you should uninstall SNAP and do an other beta test request.
If you are running the latest version, on which OS are you running SNAP? Could you also send us the messages.log file from the .snap/var/log/messages of your home directory as well as the root xml file of one of the product?
Dear @oana ,
I have had the same issue as Nicolas does here - regarding importing old file naming version: S2A_OPER_PRD_MSIL1C_PDMC_20160330T084950
Could you please assist me with more detials on how should I resolve this problem?
Please see the error messges as follows:
Thank you very much for your time in advance!
Regards,
Izzy
java.lang.IllegalStateException: The naming convention structure is invalid for input file ‘D:_2018 Research\02 Raw data\PALSAR-2_2nd order\S2A_OPER_PRD_MSIL1C_PDMC_20160615T063730_R116_V20160615T002245_20160615T002245.zip\S2A_OPER_PRD_MSIL1C_PDMC_20160615T063730_R116_V20160615T002245_20160615T002245.SAFE\S2A_OPER_MTD_SAFL1C_PDMC_20160615T063730_R116_V20160615T002245_20160615T002245.xml’
at org.esa.s2tbx.dataio.s2.metadata.AbstractS2MetadataReader.(AbstractS2MetadataReader.java:33)
at org.esa.s2tbx.dataio.s2.ortho.metadata.AbstractS2OrthoMetadataReader.(AbstractS2OrthoMetadataReader.java:19)
at org.esa.s2tbx.dataio.s2.l1c.metadata.S2L1cProductMetadataReader.(S2L1cProductMetadataReader.java:21)
at org.esa.s2tbx.dataio.s2.l1c.Sentinel2L1CProductReader.buildMetadataReader(Sentinel2L1CProductReader.java:60)
at org.esa.s2tbx.dataio.s2.l1c.Sentinel2L1CProductReader.buildMetadataReader(Sentinel2L1CProductReader.java:45)
at org.esa.s2tbx.dataio.s2.Sentinel2ProductReader.readProductNodesImpl(Sentinel2ProductReader.java:152)
at org.esa.snap.core.dataio.AbstractProductReader.readProductNodes(AbstractProductReader.java:178)
at org.esa.s2tbx.dataio.s2.ortho.Sentinel2OrthoProductReaderProxy.readProductNodes(Sentinel2OrthoProductReaderProxy.java:84)
at org.esa.snap.core.dataio.ProductIO.readProduct(ProductIO.java:179)
at org.esa.snap.rcp.actions.file.ReadProductOperation.run(ReadProductOperation.java:61)
at org.openide.util.RequestProcessor$Task.run(RequestProcessor.java:1443)
at org.netbeans.modules.openide.util.GlobalLookup.execute(GlobalLookup.java:68)
at org.openide.util.lookup.Lookups.executeWith(Lookups.java:303)
[catch] at org.openide.util.RequestProcessor$Processor.run(RequestProcessor.java:2058)
Hi,
Nicolas is not visiting this forum anymore. But maybe @Oana can have a look at this.
Thanks a lot for the headsup. Much appreicted!
Hi Marco,
I tried to get in touch with Oana for a while, unfortunately I was unable to reach out to her.
Is there someone you could kindly point me to please?
Wish you a happy holiday season and happy new year!
Regards,
Izzy
Hello! Happy New Year!
It seems the wrong Oana was tagged before 
I will have a look. I will get back to you if I need more details.
Hi Izzy,
Happy new year!
Please, could you indicate the snap version?
There is something strange with the path in the log error:
“D:_2018 Research\02 Raw data\PALSAR-2_2nd order\S2A_OPER_PRD_MSIL1C_PDMC_20160615T063730_R116_V20160615T002245_20160615T002245.zip”
Is it the correct name display by the log? Could you share the product folder path?
I would be interested to see the zip folder structure.
Hey Oana! Happy new year to you too!
Thank you very much for looking after my case. So glad we connected in 2022 =)
Please let me know if you require any further information from my end.
Best regards,
Izzy
Hi Florian,
Thanks for reaching out to me. Happy new year to you too!
Q1: Please see my SNAP version detials as follows:
- SNAP Desktop implementation version: 8.0.9
- SNAP Engine implementation version: 8.0.9
S2A_OPER_PRD_MSIL1C_PDMC.txt (411 Bytes)
Q2: Yes, that’s the correct directory for Sentinel-2 data, and I should haved sorted it out more nicely. Please see the at attahced two .txt files for more information regarding this specific dataset.
RootStructure_S2A_OPER_PRD_MSIL1C_PDMC_20160422T084127.txt (111.9 KB)
Thank you!
Best regards,
Izzy
Dear @isa_leesln ,
Since I saw the following error in your provided log, I whould need to see the zip folder structure.
(you can unzipp it and provide captures with its directories structure)
Thank you in advance.
@oana_hogoiu Thanks a lot for your prompt reply!
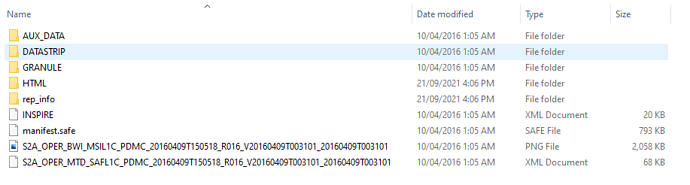
Please see the folder structure in the attached text file below. RootStructure_S2A_OPER_PRD_MSIL1C_PDMC_20160422T084127.txt (111.9 KB)
Regards,
Izzy
Nothing pops out from the structure, besides the fact that the zip doesn’t have the “.SAFE” suffix in its name.
I’ve compared with a S2A_OPER_PRD_MSIL1C_PDMC_20160527T123330_R051_V20160511T105343_20160511T105343.SAFE
product that I have, which opens ok in SNAP.
I am assuming that you have the same error if you try with the unzipped product?
I’ve PM you for sending me your product, maybe there is something else I’m missing.
Hi @oana_hogoiu ,
Thanks for checking the data. Let me recap my procedures and the problem occurred when import, hope this will make sense to you. (I probably didn’t explian myself very well in the last few threads)
Step1: import orginal zip file (drag and drop into SNAP)
Here it pops out a window asking for choose the suitable Readers 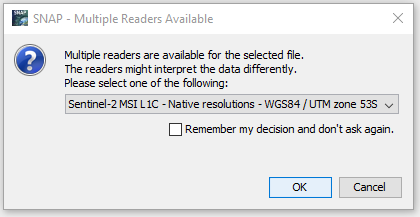
Step2: where the problem occurred and the there is no data showing up on the Product Explorer or the Display window. Only the small red cross with circle show on the right hand side corner, and then I can see the error message by clicking and expending it (savederror_msg.txt (3.0 KB)
in the txt file below).
Q: when you import the data, do you use the same way to import? If the import procedure is successful, can you see/open the image to display it in SNAP?
Thank you for your patience and help!
Regards,
Izzy
Hello
happy New Year 
I note the error messages in @isa_leesln Izzy’s error_msg.txt (Lines 13 to 16):
Caused: java.lang.RuntimeException: Provider for class javax.xml.stream.XMLInputFactory cannot be created
at javax.xml.stream.FactoryFinder.findServiceProvider(FactoryFinder.java:367)
Caused: javax.xml.stream.FactoryConfigurationError: Provider for class javax.xml.stream.XMLInputFactory cannot be created
at javax.xml.stream.FactoryFinder.findServiceProvider(FactoryFinder.java:370)
It’s not clear to me what this relates to, but I don’t believe it is an issue in data quality. I also note that the data is a slice product, which are no longer available on the SciHub; everything there is now single Tile. We cannot therefore replicate Izzy’s issue locally.
Cheers
Jan
Jan Jackson
OPT-MPC S2 Technical Manager
The XMLInputFactory error is known and does affect the reading or the processing of the data:
It has been discussed earlier here:
After update: Provider com.ctc.wstx.stax.WstxInputFactory not found - snap / Problem Reports - STEP Forum (esa.int)
Hello @isa_leesln @marpet @oana_hogoiu
I have installed SNAP 2_2, and SNAP 3.0. Neither of them will open these Slice products (I was able to get the product from https://copernicus.nci.org.au/sara.client/#/home eventually; at 7GB-plus it takes a considerable time to download).
The Slice will also not open in SNAP 8. It returns a message of:
“java.lang.IllegalStateException: The naming convention structure is invalid for input file” and attempting to open the XML of the product (or individual Tiles) returns the message:
“No appropriate product reader found.”
However, the naming convention structure is compliant with the Product Specification Document of these products (PSD 12).
I note that individual Image Bands (JP2 format) from the individual Tiles contained in the Slice product can be visualised in SNAP.
I cannot see an issue with the product format, or structure. I would recommend that the Compact Naming Convention Tiles available from the SciHub be used instead of these old format products.
Cheers
Jan
OPT-MPC S2 Technical Manager
Hi @Jan,
Thank you very much for looking into the issue. The actual data can be found via https://copernicus.nci.org.au/sara.client/#/home, where has the archived data all the way back to 2014. Hope this could help, if otherwise I can provide the data file with a share link.
Please let me know what works for you.
Regards,
Izzy
Hi @isa_leesln Izzy
I found the data, but - barring the fact that it won’t open in the various version f SNAP I’ve tried - I couldn’t see any issues with it. I can open the individual Band IMG data (JP2 format) in QGIS.
I checked it against another slice product, and the construction of the metadata, and the filename convention appears OK. Thus, my recommendation to use single Tile production from this date.
Cheers
Jan