I am trying to write a code that goes through the RADAR pre-processing steps to look at agricultural fields and have based my code off of this one and this is what I have:
import datetime
import time
import sys
sys.path.append(r'C:\Users\EM\.snap\snap-python')
from snappy import ProductIO
from snappy import HashMap
import os, gc
from snappy import GPF
def apply_orbit_file(source):
print('Apply orbit file...')
parameters = HashMap()
parameters.put('Apply-Orbit-File', True)
output = GPF.createProduct('Apply-Orbit-File', parameters, source)
return output
def remove_grd_border_noise(source, polarization, pols):
print('GRD border noise removal...')
parameters = HashMap()
if polarization == 'DH':
parameters.put('sourceBands', 'Intensity_HH,Intensity_HV')
elif polarization == 'DV':
parameters.put('sourceBands', 'Intensity_VH,Intensity_VV')
elif polarization == 'SH' or polarization == 'HH':
parameters.put('sourceBands', 'Intensity_HH')
elif polarization == 'SV':
parameters.put('sourceBands', 'Intensity_VV')
else:
print("different polarization!")
parameters.put('selectedPolarisations', pols)
output = GPF.createProduct('Remove-GRD-Border-Noise', parameters, source)
return output
def thermal_noise_removal(source):
print('Thermal noise removal...')
parameters = HashMap()
parameters.put('removeThermalNoise', True)
output = GPF.createProduct('ThermalNoiseRemoval', parameters, source)
return output
def calibration(source, polarization, pols):
print('Calibration...')
parameters = HashMap()
parameters.put('outputSigmaBand', True)
if polarization == 'DH':
parameters.put('sourceBands', 'Intensity_HH,Intensity_HV')
elif polarization == 'DV':
parameters.put('sourceBands', 'Intensity_VH,Intensity_VV')
elif polarization == 'SH' or polarization == 'HH':
parameters.put('sourceBands', 'Intensity_HH')
elif polarization == 'SV':
parameters.put('sourceBands', 'Intensity_VV')
else:
print("different polarization!")
parameters.put('selectedPolarisations', pols)
parameters.put('outputImageScaleInDb', False)
output = GPF.createProduct("Calibration", parameters, source)
return output
def speckle_filtering(source):
print('Speckle filtering...')
parameters = HashMap()
parameters.put('filter', 'Lee')
parameters.put('filterSizeX', 3)
parameters.put('filterSizeY', 3)
output = GPF.createProduct('Speckle-Filter', parameters, source)
return output
def terrain_correction(source):
print('Terrain correction...')
parameters = HashMap()
parameters.put('demName', 'SRTM 3Sec')
parameters.put('imgResamplingMethod', 'NEAREST_NEIGHBOUR')
parameters.put('saveProjectedLocalIncidenceAngle', True)
parameters.put('saveSelectedSourceBand', True)
output = GPF.createProduct('Terrain-Correction', parameters, source)
return output
def main():
## All Sentinel-1 data sub folders are located within a super folder (make sure the data is already unzipped and each sub folder name ends with '.SAFE'):
path = r'D:\RD\radar\pythontest\in'
outpath = r'D:\RD\radar\pythontest\out'
if not os.path.exists(outpath):
os.makedirs(outpath)
for folder in os.listdir(path):
gc.enable()
gc.collect()
sentinel_1 = ProductIO.readProduct(path + "\\" + folder + "\\manifest.safe")
print(sentinel_1)
loopstarttime=str(datetime.datetime.now())
print('Start time:', loopstarttime)
start_time = time.time()
## Extract mode, product type, and polarizations from filename
modestamp = folder.split("_")[1]
productstamp = folder.split("_")[2]
polstamp = folder.split("_")[3]
polarization = polstamp[2:4]
if polarization == 'DV':
pols = 'VH,VV'
elif polarization == 'DH':
pols = 'HH,HV'
elif polarization == 'SH' or polarization == 'HH':
pols = 'HH'
elif polarization == 'SV':
pols = 'VV'
else:
print("Polarization error!")
if modestamp == 'IW' and productstamp == 'GRDH':
# Start preprocessing:
applyorbit = apply_orbit_file(sentinel_1)
grd_removed = remove_grd_border_noise(applyorbit, polarization, pols)
thermal_removed = thermal_noise_removal(grd_removed)
calibrated = calibration(thermal_removed, polarization, pols)
speck_filtered = speckle_filtering(calibrated)
down_tercorrected = terrain_correction(speck_filtered)
del applyorbit
del grd_removed
del thermal_removed
del calibrated
del speck_filtered
# save final product
#TODO: figure out how to get the separate polarizations
print("Writing...")
ProductIO.writeProduct(down_tercorrected, outpath + '\\' + folder[:-5], 'GeoTIFF-BigTIFF')
del down_tercorrected
print('Done.')
sentinel_1.dispose()
sentinel_1.closeIO()
print("--- %s seconds ---" % (time.time() - start_time))
else:
print("Different product type found...need IW GRDH.")
if __name__== "__main__":
main()
My output won’t load into QGIS, but when I put it in snap I see the tif and the bands:
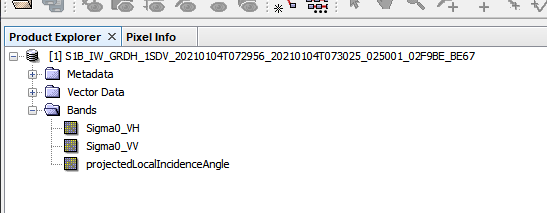
but when I try to view the sigma0 bands I get this error:
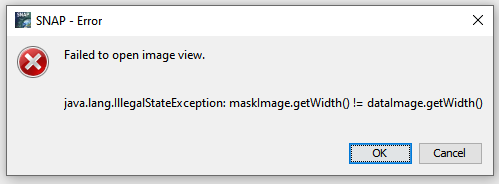
it seems like there must be something wrong with the metadata written to the image and it doesn’t know the proper raster size?
I must be missing a step but I am not sure where.