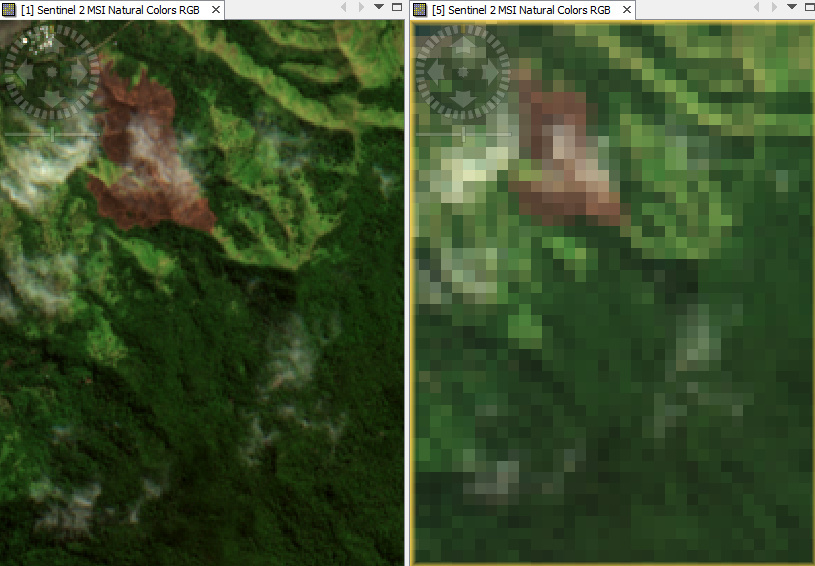
Yes. I’ve also tried running the tool from the command line. Looking at the outputs I find that in the GRANULE folder with the IMG_DATA directory I only have R20 and R60 folders:

The R20 folder only holds what looks like an intermediary file:
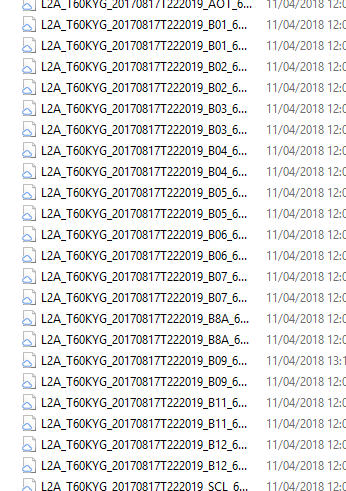
Whereas the R60 folder holds all bands at 60m:
Running the tool from the cli afforded me the opportunity to see the readout and something does go wrong. Here’s the output from the cmd window:
Traceback (most recent call last):
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\lib\multiprocessing\process.py”, line 258, in _bootstrap
self.run()
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\Lib\site-packages\sen2cor\L2A_ProcessTile.py”, line 124, in run
if not self.process_20():
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\Lib\site-packages\sen2cor\L2A_ProcessTile.py”, line 158, in process_20
return self.process()
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\Lib\site-packages\sen2cor\L2A_ProcessTile.py”, line 223, in process
if ac.process() == False:
File “L2A_AtmCorr.py”, line 3163, in L2A_AtmCorr.L2A_AtmCorr.process (L2A_AtmC
orr.c:73951)
File “L2A_AtmCorr.py”, line 7710, in L2A_AtmCorr.L2A_AtmCorr.rho_retrieval_ste
p1 (L2A_AtmCorr.c:204885)
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\Lib\site-packages\sen2cor\L2A_Tables.py”, line 2598, in getDataType
h5file = open_file(self._ImageDataBase, mode=‘r’)
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\lib\site-packages\tables\file.py”, line 320, in open_file
return File(filename, mode, title, root_uep, filters, **kwargs)
File “C:\myInstallLocation\Sen2Cor-02.05.05-win64\Sen2Cor-02.05.05-win6
4\lib\site-packages\tables\file.py”, line 783, in init
self._g_new(filename, mode, **params)
File “tables\hdf5extension.pyx”, line 487, in tables.hdf5extension.File._g_new
(tables\hdf5extension.c:5943)
HDF5ExtError: HDF5 error back trace
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5F.c”, line 604, in H5Fopen
unable to open file
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5Fint.c”, line 1087, in H5F_open
unable to read superblock
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5Fsuper.c”, line 294, in H5F_super_read
unable to load superblock
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5AC.c”, line 1262, in H5AC_protect
H5C_protect() failed.
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5C.c”, line 3574, in H5C_protect
can’t load entry
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5C.c”, line 7954, in H5C_load_entry
unable to load entry
File “C:\sen2cor\deps\hdf5-1.8.18\src\H5Fsuper_cache.c”, line 476, in H5F_sblo
ck_load
truncated file: eof = 389409061, sblock->base_addr = 0, stored_eoa = 4634678
78
End of HDF5 error back trace
This would explain why when the tool is run in SNAP only a 60m output can be viewed. However, I don’t know what is causing the issue.
Next I will try removing the standalone install of sen2cor, resetting my paths, uninstalling the plugin from SNAP, reinstalling the plugin to SNAP and then insallting the bundle through the SNAP interface. I’ll report back what I find.