Hello
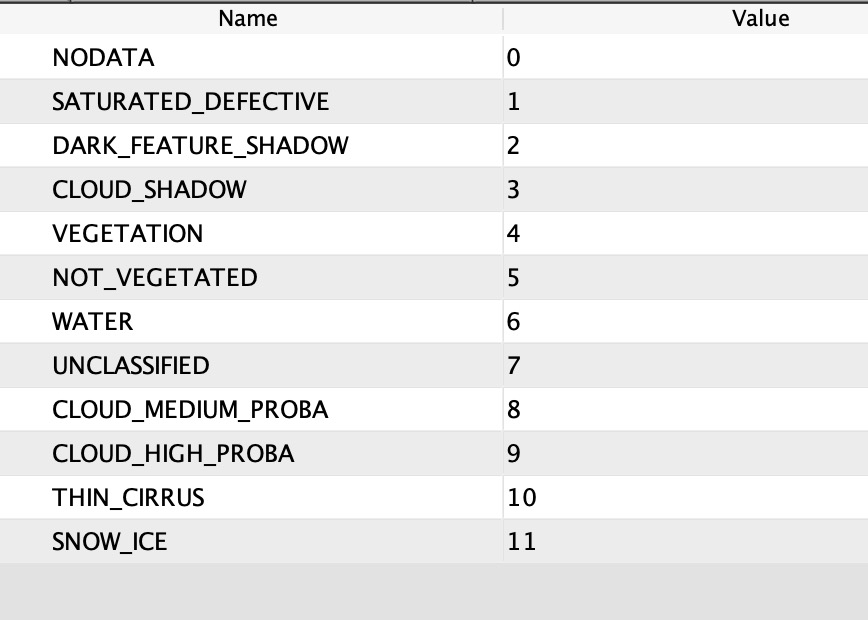
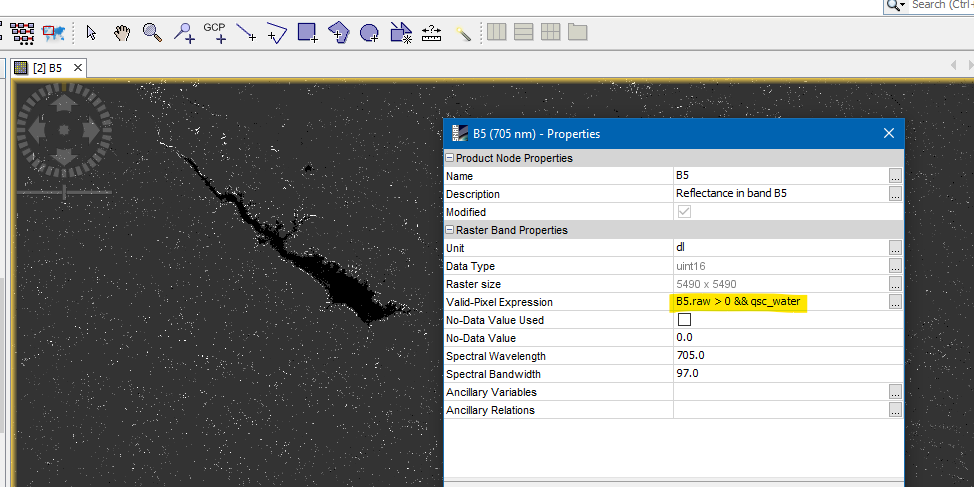
I have followed the septs you indicated me in your last post and tried in S2A_MSIL2A product to use the “quality_scene_classification” for masking and worked really well for water
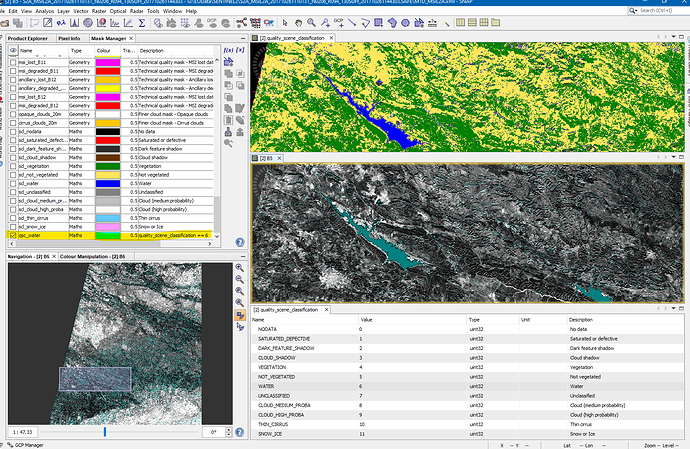
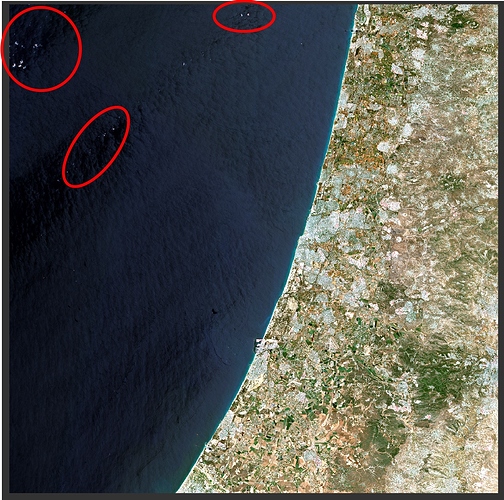
However when tried to mask clouds I find the following issue, clouds in the sea are clearly visible and detected
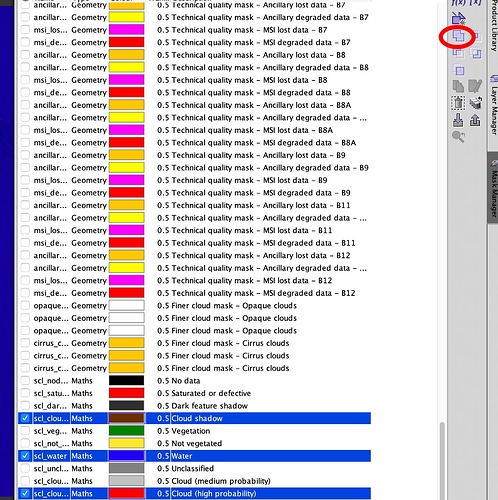
while clouds are in general visible in RGB image in land and sea, although, for clouds on land when I zoom in an extremely large number of pixels (which in the RGB don’t appear to have any visible clouds by eye), show to be with clouds cover all over the image taking a very large number of the total pixels in the whole image. Thus, it captured my attention the very large and important amount of pixels the mask indicates to be with clouds. I used the "cloud (high probability) mask. While it does not look to be any clouds in the RGB image in the the mask such large number of pixels appear in the cloud (high probability) mask… In RGB as you can see besides the could in the sea there does not appear to be major clouds?
Why could this be happening ?
How I can get a reliable and more appropriate mask for clouds and clouds shadows on land from “quality_scene_classification”?
My objective is to make a union mask (which I actually could do) of the water, clouds and clouds shadow aiming at using such mask to set null those pixels in the image.
How can I use the mask to set null pixels in the image?
For instance to set null the mask resulting from the union of water, clouds and shadows?
Is there a setnull() function in SNAP such as in ArcGIS?
If exists how could be the syntax for setting null in the image the mask values?
Then, finally with an appropriate making of all water, cloud and shadow, pixels my preprocessing steps will finish to start using the image for research purposes.
For that I would need to extract as GeoTIFF several bands (not all of them) from the product, this I think can be done by separating the bands from the product using subset function, and only then export as GeoTIFF is this correct?
After that for completing the process I think I have to multiply the GeoTIFF times the scaling factor if is not in reflectance between 0 and 1 as in other post in this forum was discussed.
Looking forward to your response.
Thanks a lot for your help.
Kind regards,
Gabriel