Hello,
I created a graph using the graph builder which I edit iteratively using python code. I am batch-processing Sentinel-3 SLSTR data. Part of the graph includes a reprojection to WGS84 geofraphic and a subset process based on wkt. Everything worked great and I processed hundreds of images. Since yesterday, without me changing anything, suddenly the reprojected and subsetted results appear in west Africa around (0,0).
When I import the graph to SNAP, add the Read and Write nodes and run manually, everything works fine. I have no idea what else to try. Please help!
I think it has something to do with the geoRegion parameter in the subset node, but I m not sure.
I am attaching the the graph I used to this post.
Thank you
slstr_preprocess_cal_gpf_EASY.xml (3.9 KB)
Yes, your assumption is right.
The region and the geoRegion create a target product with zero size.
Maybe something wents wrong when you alter the graph in python?
But actually it should not work in SNAP either.
Dear Marco,
Thank you for the answer!
However why is it a zero-sized region? There is a difference in the coordinates of the rectangle points. When I use a wkt plotter it seems to plot the georegion area correctly.
I also noticed that the resulting raster does not implement the geographic scale correctly and each pixel is treated as a 1 x 1 degree cell.
I uploaded the graph I used after I updated the geoRegion and a snapshot of the resulting raster.
Thank you for your help!
Or did I misunderstood something?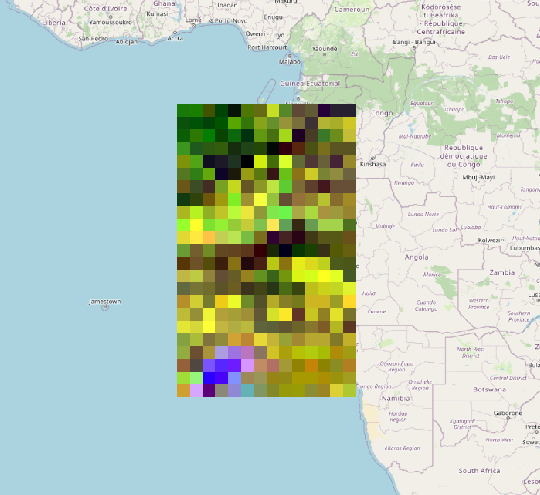 slstr_preprocess_cal_gpf_EASY.xml (3.9 KB)
slstr_preprocess_cal_gpf_EASY.xml (3.9 KB)
Oh, sorry. For me, all coordinates looked the same. Didn’t looked carefully enough.
In this case, I understand it neither.
Dear Marco,
Could you please point me in the right direction for the solution?
I rechecked everything in the python code and it seems to be fine.
The main curiosity -
- I recreated a graph in the graph builder GUI, ran it and made sure it works.
- Exported it to a xml file, removed the Read and Write nodes
- Ran the edited xml graph from the terminal with gpt
- The resulting raster is now located in the ocean next to west Africa again.
I have no more ideas 
Can it be a newly introduced bug?
Thank you
slstr_preprocess_cal_gpf_EASY.xml (3.9 KB)
I’m not sure what to suggest.
Do you the latest updates installed?
You could make the graph shorter. Maybe start with Rad2Refl ans Resample and take a look at the result. If okay, then add another node. This way we see, which one causes the issue.
I am confident it is the “Reproject” node. Each other node that was tested individually seems to work correctly. Attached is the shortened graph I used:
slstr_preprocess_cal_gpf_MUIR_2_ed.xml (2.3 KB)
and it’s result can be seen in the image

I use NSAP 7.0
Now, I think it has something to do with the installation of SNAP.
If i run exatly the same graph on the same image on my personal laptop everything works fine.
The working station and my working laptop have the same operating system, and the same version of SNAP.
I will try to reinstall SNAP and gpt on the working station and update with the results
EDIT: It did not work.
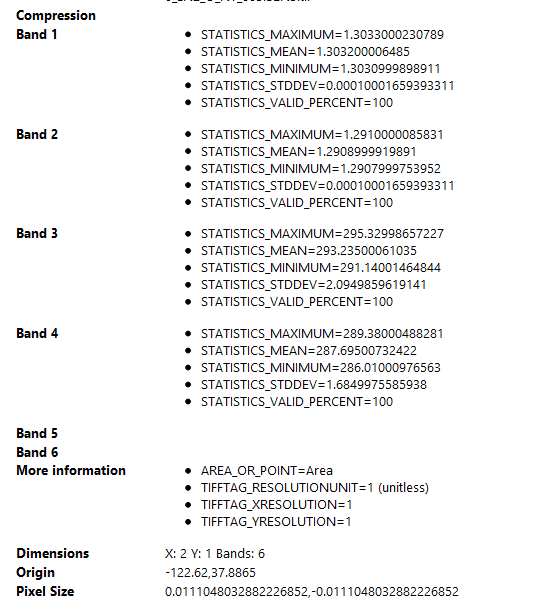
These are the differences between the properties of the same image processed with the same graph on two different work stations:
Work station (incorrect)
Home work station (Correct)
SOLUTION: replaced all files in snap/bin directory on the working station with files from the same directory on my laptop. Everything works!