Hi all,
I’m currently working with LAI, FPAR and FCOVER data retrieved from the Biophisycal operator. I ran the module from command line using and R script to automate the process.
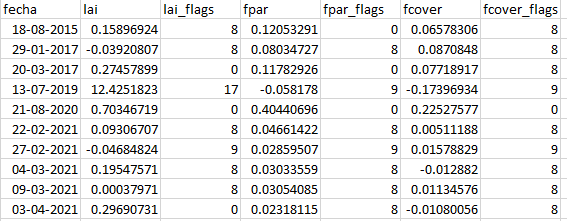
When i check the output data it looks good, for example the values and ranges of the variables.
My question is regarding the quality flags. Reading the manual (https://step.esa.int/docs/extra/ATBD_S2ToolBox_L2B_V1.1.pdf) it looks that the QA flags could take values from 0 to 4 (3 bits) but i have strange values (image attached), it seems that the 0 value is ok, the 1 value is fair ok (input out of range) but the values above 4 are bad quality data. The majority of the QA data is beetween 0 and 1 but i think that if may be and error in the computing of the flags, maybe it could be a error in the computing of the LAI, FPAR and FCOVER products also.
Someone have had this error?
Best regards,