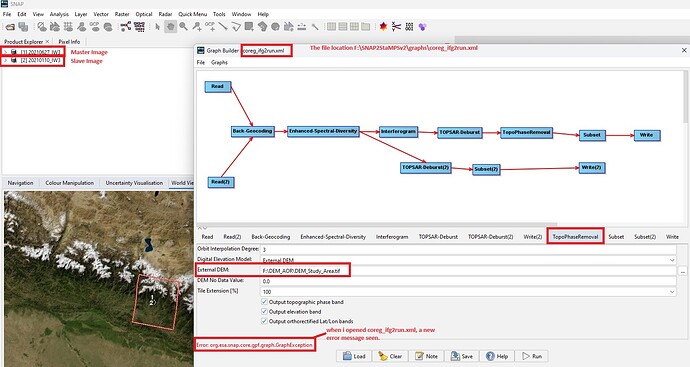
@mdelgado : I am sorry sir for missunderstanding the External DEM you mean. Now I opened coreg_ifg2run.xml file and checked the DEM file path. Please check it.
Also, Please find the DEM file of my AOR.
https://drive.google.com/drive/folders/1X-muWgBiEWv2EG1Kd2-6-lqhsaaRMDoN?usp=drive_link
Similarly, I am attaching the ROA shape file.
Try to use the Ext DEM only for the TopoPhaseRemoval manually… and let me know.
Btw… are you using snap 9?
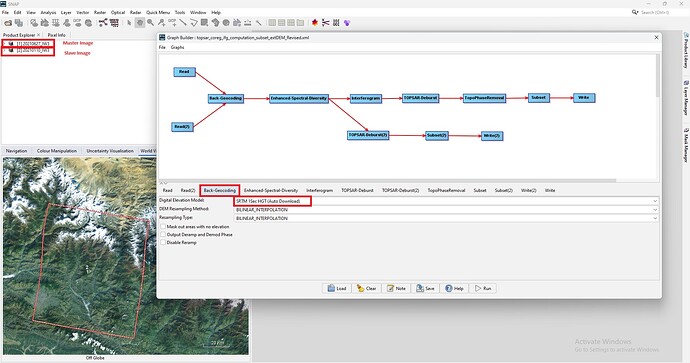
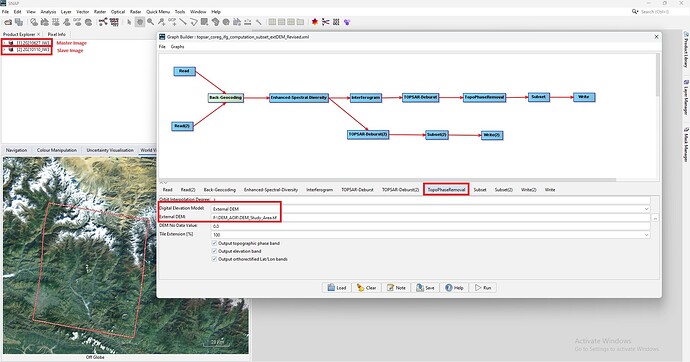
@mdelgado: Dear Sir, I try to run coregistration and interferometric generation manually using one master and one slave image. This time i tried only using external DEM at Topographica Phase Removal as shown below.
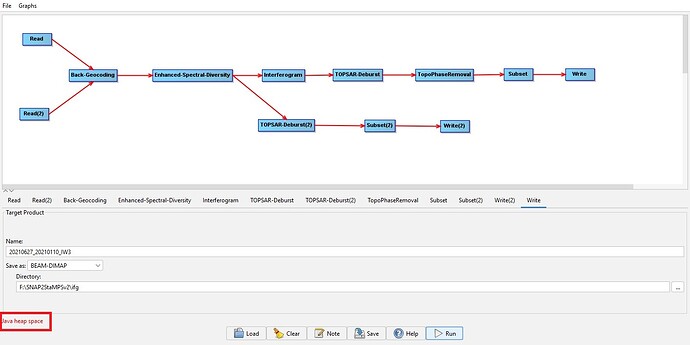
Intially it starts execution and after 5% of progress the error “Java heap space” comes as below and the software shutdown automatically.
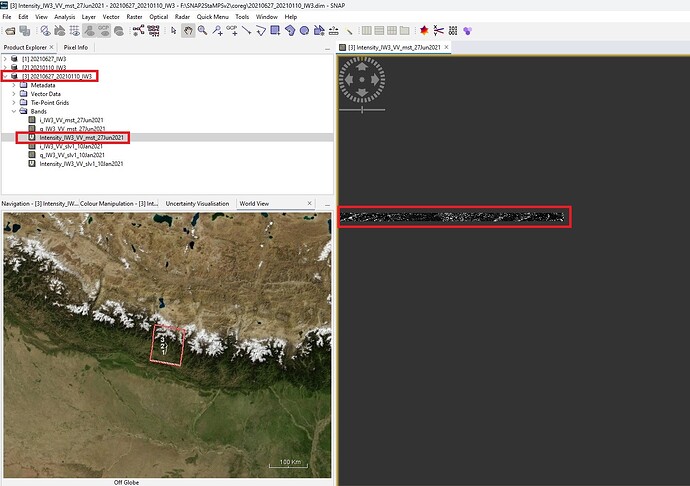
But, When i try to open the “coreg” image it shows as below.
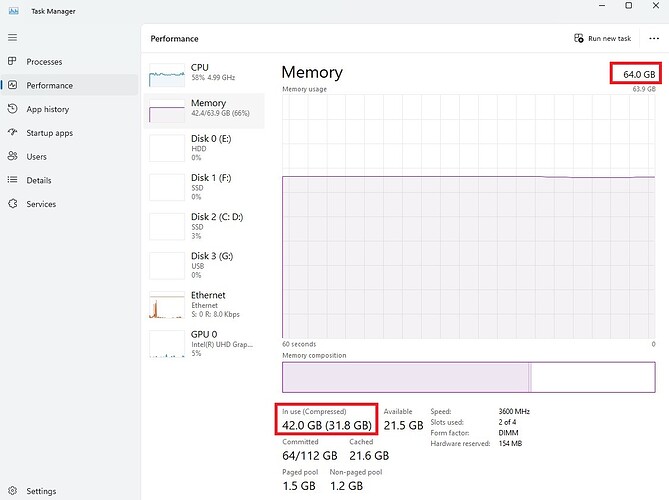
For yours kind information, I am using SNAP 10.0 version (latest) in my Windows 11. When processing SNAP only 8.4 GB RAM is utilized out of 32 GB. Please find the .xml file i have revised this time.
topsar_coreg_ifg_computation_subset_extDEM_Revised.xml (7.6 KB)
i don’t know what “Java heap space” error message displayed for.
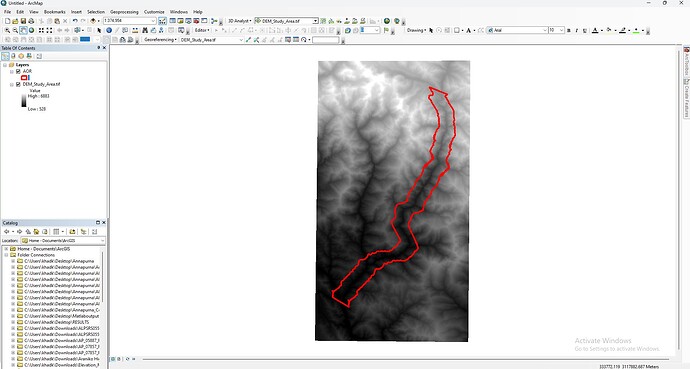
@mdelgado : For your kind information, I am attaching the DEM image of my study area as below.
Is it error due to the size of DEM? I still can’t figure out the problem? Looking forward for yours suggestion.
@mdelgado: Dear Sir, Since i observed “Java heap space” error, i update my RAM capacity and re-run coregistration & interferometric generation manually with single master and single slave. This time again i use external DEM at topographic phase removal only. I am using SNAP 10.0. But, this time also the progess is made upto 5% and SNAP is closed authomatically after running for 5 hrs.
Please check .xml file i have used.
topsar_coreg_ifg_computation_subset_extDEM_Revised.xml (7.6 KB)
I am looking for yours suggestion.
Java Heap Error usually means that you shall increase the memory of the JVM with the -Xmx parameter, on the SNAP configuration.
I am wondering if your AOI is too large or so, but with the proper configuration all shall run.
Let me know
Please send me 2 filenames of the S1 SLCs used so I can try to replicate/solve your issue.
Best
@mdelgado :Thank You sir for yours response. Here is the SLC file attached.
20210627_IW3.dim (2.3 MB)
20210110_IW3.dim (2.3 MB)
I used 20210627 as master and 20210110 as slave image earlier.
I expected this 
S1A_IW_SLC__1SDV_20210627T001133_20210627T001200_038518_048BA5_7DF2
S1A_IW_SLC__1SDV_20210110T001130_20210110T001157_036068_043A35_E57F
Thanks. I will download it and give a try.
I will keep you posted
@mdelgado : I miss understood Sir, I send the SLC image after selecting study area by choosing sub-swath and brust. Here is the image as per your interest. Since, the size of image is more than 4 GB, I am sharing you the google drive link.
I used sub-swath “IW3” and burst “1 to 4” for the above images.
@mdelgado : Hello Sir, I hope you could download the SLC images from Google Drive link i have shared.
Diwakar
Hi! I did not managed from you GDrive links, but I got them anyway from ASF. However, I keep getting error with the ExternalDEM, so let me check some more possibilities.
The main issue lies on the fact that your external DEM is not on WGS84, but on Pseudo WGS84 (with meter units instead of degrees).
Additionally, note that your DEM had zero data values as 32767 and not 0.0. Those 2 things were causing your issues.
on the snap2stamps Github repo site, there was already a warning exactly referring to this:
" Warning : If the user wants to use its own External DEM, please ensure it is in geotiff format with no data values as 0.0 and preprocessed using egm84 ellipsoid. Otherwise, ensure to adapt the parameters in the xml graphs accordingly."
So by taking care of what written some issues could had been avoided.
Please remember that you need to reproject your DEM into wgs84 projection to use the externalDEM.
I hope this helps you to avoid your error
@mdelgado Thank you sir, after necessary correction, i will get back to you.