If you want it you can change it if course. You can do the same looking at the phase.
Personally I prefer to save time and disk space, but you may prefer the way you want.
Feel free to change that option on the xml graph.
If you want it you can change it if course. You can do the same looking at the phase.
Personally I prefer to save time and disk space, but you may prefer the way you want.
Feel free to change that option on the xml graph.
Thank you!
It is a nice reference image to use since looking at phase, unwrapped phase and erros can be quite challenging for new people in the insar realm.
And going to multiple iterations of step 6 and 7 to get the good results requires alot of knowledge of how the images should look.
making the coherence maps was luckily no problem!
now i have the coherence maps i see the problems in my processing, what looks like temporal de correlation is insane:
20180803 master 20180610 slave (very bad coherence)
20180803 master 20180908 slave (good coherence over the land area)
my AOI is in a permafrost area and 2018 was cold so probably snow coverage could be the problem here but alot of these bad areas are picked up by stamps as ps points.
i already have only 20 images between 201806 and 201810 so data reduction is hard, i do have images for 2016 aswell (9 images) but i am interested in the seasonal subsidence change between 2016 and 2018 due to an event in 2017 so combining the 2 years is not really an option right?
Hi Stoorm,
I am having a similar problem to you. I am using Ubuntu 16.04 with python 2.7, and the following error when running splitting_slaves.py. Did you ever find a solution to this problem?
Update - I think I may solved this by reading a previous posts: I needed to specify the full path of the gpt file in project.conf.
Second, My AOI is covering left and right paths of S1.
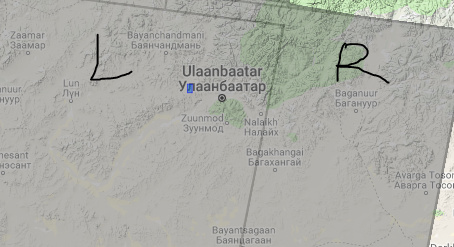
Therefore, i have collected 36 images from each side.Now i am started process left side images. Next, will start right side. Up till now, processing steps working fine.
So, can i merge left and right sides results in stamps?
If the l and r you refer to are different path/frame combinations you can combine this results after the stamps-processing but not before.
Thanks for your URL you shared with us. It helpful for me.
Dear Jose Manuel,
Thank you very much for presenting such a fantastic tool!
I am a new user of snap2stamps (also a fresher of SNAP-Stamps workflow). Now, I had succeded in running the “splitting_slaves.py” script. But I found it is very slow to run the “coreg_ifg_topsar.py” script. It did not finish the processing of the first slave file after 12 hours of running!
Can you give me some cues?
This project has 90 slaves. Would this be the problem?
Or, is it related to the CPU/Memory and the configuration of my PC?
On my PC, it spent about 170 seconds for each slave in the “splitting_slaves.py” step.
Thank you very much!
I am almost certain this due to your area being out of reach of the DEM you are using.
i have had the same problem and ended up using TAN-DEM inplace of the SRTM tiles.
in this post you can read how to change the DEM parameters in SNAP2StaMPS.
Goodluck!
@Gijs
Thank you very much! I will try the solution that you suggested.
Thanks for this free package. Nice.
@Gijs @mdelgado
Dear Gijs (and any other guys concerning my problem). I am sorry to borther you again. Unfortunately, I tried the solution that you suggested but still got very very slow processing in the run of the “coreg_ifg_topsar.py” script in snap2stamps. I used an external tif DEM, a mosaiced one which is large enough to cover the whole SAR images. I had tried both mosaiced SRTM and TAN-DEM.
I run the “coreg_ifg_topsar.py” script at 14:09 on 2019/05/27, and got some file in the coreg and ifg folders at around 03:00 O’clock on 2019/05/28. Now, it is 08:30 on 2019/05/29, but the processing is still sticking at the first slave image, with nothing new appeared in the coreg and ifg folders.
I upload my “coreg_ifg_computation.xml” , “project.conf”, and some screenshots of the coreg and ifg folders. Can you help me out of this?
Thank you very much!
coreg_ifg_computation.xml (5.7 KB)
project.conf (660 Bytes)
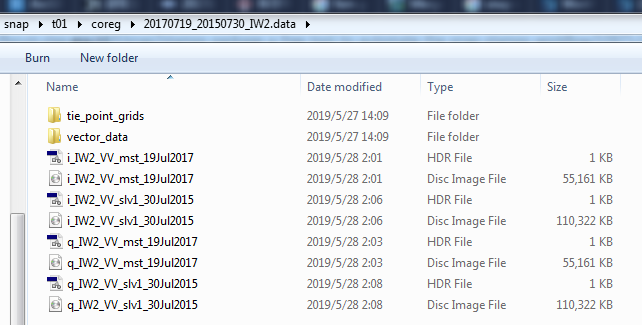
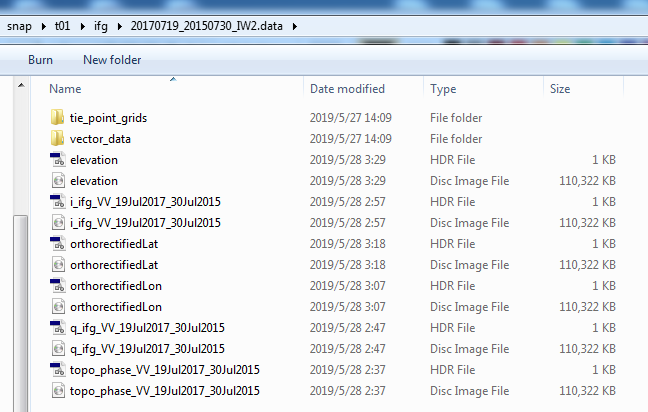
When you say you mosaiced SRTM and TanDEM-X, does that mean that your data ranges over several degrees?
Is it projected to WGS84 by the way?
@ABraun
Dear ABraun. Yes. The range of my mosaiced SRTM and TanDEM is 114°E ~ 120°E and 32°N ~ 37°N。And, their coordinate system is GCS_WGS_1984.
I just wonder how the data can cover such large range over (500 km?) and i and q are still 110 MB only. Usually, the size of the file is the final at an early stage of processing so they won’t become any larger.
I am not sure about the reason for the smal size of output img files. Maybe it is because I applied an AOI during the processing?
One thing is sure:
Snap is processing something it cant process but doesnt not output an error.
I had this when my DEM didnt cover the area but you said that wasnt the case.
maybe you are accidentally combining descending and ascending data? or strips that do not overlap perfectly.
is your DEM in unprojected WGS_84 and saved as a geotiff.tif
Unexpectedly, the processing of the first slave image finished just now, at around 16:10 on 2019/05/29. So, it spent 50 hours! I don’t know whether the output files are correct. I attach screenshots here too.
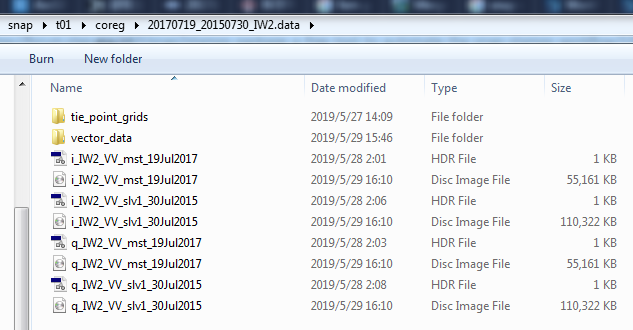
However, even if the outputs are correct. It is unaccpetable to spend 50 hours on a single slave image!
The mosaic DEM I used is in geotiff.tif format, and in unprojected WGS_84.
All the 91 SAR iamges have the same path number (ascending), but with different frame numbers. Although data with different frames are used, all of them cover my AOI. Might different frame numbers be the reason?
I have used different frame numbers aswell with succes so this should not be the problem but it could be, can you tell us wich track and path/frame combinations you are using?
Also an image of the ifg and coregistration could help!
Are you also sure you have set the SNAP processing parameters correctly? xmx-max to min of 10 gb or something.