Yes, using SNAP (manually) you can select any bursts.
open the master image in SNAP, and go to S-1 TOPS Split windows, we have the option to select the sub-swath as well as Bursts.
Based on your master image all the slave images will process.
Yes, using SNAP (manually) you can select any bursts.
open the master image in SNAP, and go to S-1 TOPS Split windows, we have the option to select the sub-swath as well as Bursts.
Based on your master image all the slave images will process.
I did, but when my master image had 2 bursts, slave images remained in all IW as a result of splitting_slaves operation in snap2stamps
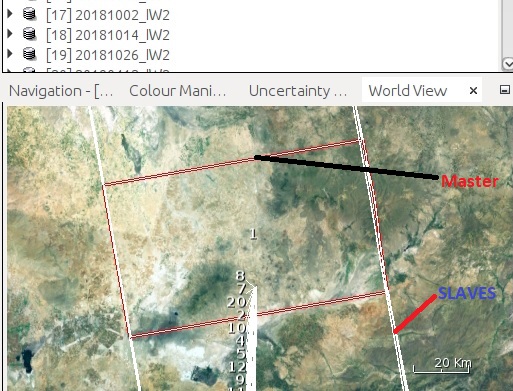
My transactions,
I applied split (2 burst) and orbit operations to the master image, respectively.
Then I did slaves_prep.py and splitting_slaves.py.
My master image has 2 bursts while my slaves images remain all IW.
While generating the interferograms (master-slave), it will definitely contain only two brusts.
Better to process all the steps, then check the final result.
yes interferograms are formed according to AOI but not in split process ?
That’s what I’m thinking.
slaves are split only based on the specified IW and not the specified AOI.
This is that way to avoid any possible miss-alignment between master and slaves ( I have seen several cases when this happened). It may not be optimal in terms of disk space etc, but ensures that the AOI is totally covered by the slaves
The final ifgs are subsetted using the AOI specified on the project.conf file.
I hope this clarifies the question.
Hi,
I am getting the following error in my 5 images, what should I do?
SNAP STDOUT:INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 2.2.3 found on system. JNI driver will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
Executing processing graph
INFO: org.hsqldb.persist.Logger: dataFileCache open start
…10%…20%…INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Shifts written to file: /home/rs/.snap/var/log/IW2_range_shifts.json
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 1/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 2/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 3/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 4/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 5/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 6/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 7/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 8/8
WARNING: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: NetworkESD (azimuth shift): arc = IW2_VV_mst_02Sep2019_IW2_VV_slv1_16Jul2019 overlap area = 0, weight for this overlap is 0.0
WARNING: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: NetworkESD (azimuth shift): arc = IW2_VV_mst_02Sep2019_IW2_VV_slv1_16Jul2019 overlap area = 1, weight for this overlap is 0.0
WARNING: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: NetworkESD (azimuth shift): arc = IW2_VV_mst_02Sep2019_IW2_VV_slv1_16Jul2019 overlap area = 2, weight for this overlap is 0.0
WARNING: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: NetworkESD (azimuth shift): arc = IW2_VV_mst_02Sep2019_IW2_VV_slv1_16Jul2019 overlap area = 3, weight for this overlap is 0.0
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Shifts written to file: /home/rs/.snap/var/log/IW2_azimuth_shifts.json
…30%…40%…50%…60%…70%…80%…90% done.
– org.jblas INFO Deleting /tmp/jblas5290024103253089492/libquadmath-0.so
– org.jblas INFO Deleting /tmp/jblas5290024103253089492/libjblas.so
– org.jblas INFO Deleting /tmp/jblas5290024103253089492/libgfortran-4.so
– org.jblas INFO Deleting /tmp/jblas5290024103253089492/libjblas_arch_flavor.so
– org.jblas INFO Deleting /tmp/jblas5290024103253089492
But I didn’t get this error on other 90 images. @mdelgado
I believe is not an error but I suggest you to check the IFG as it may happen that the ESD on that pair had not worked properly.
Thank you so much @mdelgado
Hello
I’m trying to split and update the orbit information for each of the slave images, but unfortunately, I encountered an error as is evident in the screen
If anyone has had the same problem or has an idea of how to solve it, thanks.
thanks
Looking at the command is clear you let the fields CACHE and CPU from the project.conf empty!
That was an easy error you made
thank you for your answer
it works fine now
Thank you very much.
I still have a problem.
I try to do a master-slave coregistration and generate interferograms by running the command: python coreg_ifg_topsar.py project.conf
This starts to work fine (screen 1), but it doesn’t progress.
I mentioned the appearance of two folds of interferograms and the coregistration containing a single interferogram, but when I tried to open it in Snap, it showed me an error(screen 2).
thanks
It is much better to post text error messages as text so they can be found by searches (e.g., by the next person who encounters the same error) and because it is easier to quote the text. You show a “file not found” error. You should verify that the file really doesn’t exist and check for typographical errors or permission problems.
You have both backslashes and frontslashes in the same paths, which cannot work.
yes, absolutely right, thank you