To select the full area, just go to the below webpage, and select your area of interest. just check in the copy&paste tab and change MARC to CSV. Just copy it to the project.config file.
“Full burst processing allowed. No bounding box definition needed. New option of no subseting after coregistration or interferogram to be performed.”
or you can set :
LONMIN=-180
LONMAX=180
LATMIN=-90
LATMAX=90
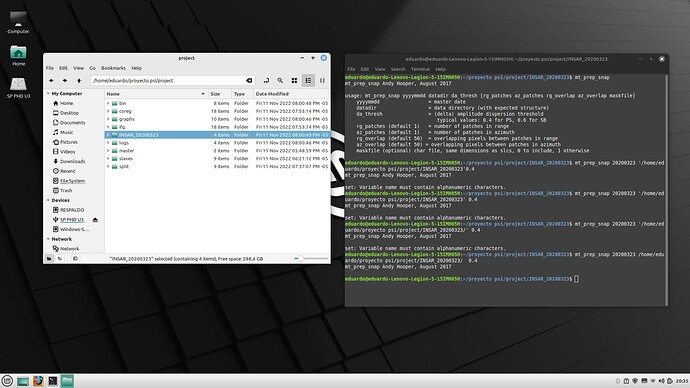
hello everyone, I have a problem related to the final process before jumping to Matlab. As you can see when i try to set mt_prep_snap I get this message of variable name must contain alphanumeric characters. Does anyone knows why this is happening? any help is accepted.
Just remove the spacing in between the proyecto psi, then try.
thank you, I did it but now when i run the script it gives me this error :C did you know why this could be happening?
Try this maximum it will solve your error.
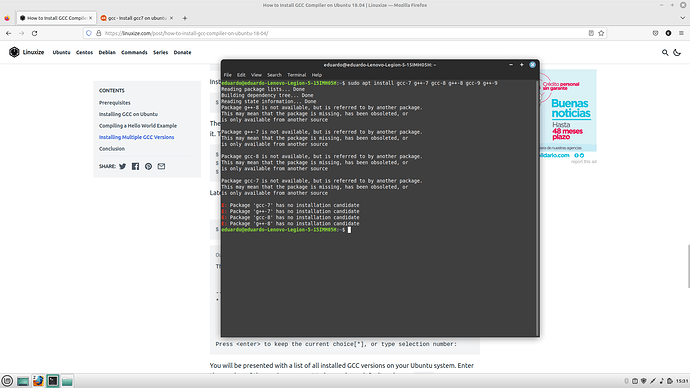
actually I tried but it gives me this error at installing the compilers, I’m starting to think that maybe is because of the version (latest mint version), do you think that running ubuntu 20.4 or 18.04 is better to do the processing?
thank you in advantage.
Ubuntu 18.04 is best.
Thank you so much, I already installed it and sorry to ask you but which version of snap and Matlab it’s suitable for Ubuntu 18.4? I got the last release but I don’t know if this would cause problems later
the latest versions are best (SNAP, StaMPS, and Matlab).
i tried to full processing for the 4 burst but your advice dosent work for me do you know another way for full IW processing.
Good night, Mr. delgado, sorry if I bother you, but I want to know if there is still no estimated date for the big update of snap2stamps? since I have had problems due to the obsolescence of certain libraries and inconveniences with unix systems related to using old versions that still support those libraries. I appreciate your further response.
hi,
is this an error?
SNAP STDOUT:INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 2.2.3 found on system. JNI driver will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
Executing processing graph
INFO: org.hsqldb.persist.Logger: dataFileCache open start
…10%…20%…INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Shifts written to file: /home/rs/.snap/var/log/IW2_range_shifts.json
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 1/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 2/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 3/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 4/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 5/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 6/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 7/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Estimating azimuth offset for blocks in overlap: 8/8
INFO: org.esa.s1tbx.sentinel1.gpf.SpectralDiversityOp: Shifts written to file: /home/rs/.snap/var/log/IW2_azimuth_shifts.json
…31%…42%…53%…63%…75%…85%… done.
– org.jblas INFO Deleting /tmp/jblas949692298280708135/libquadmath-0.so
– org.jblas INFO Deleting /tmp/jblas949692298280708135/libjblas_arch_flavor.so
– org.jblas INFO Deleting /tmp/jblas949692298280708135/libgfortran-4.so
– org.jblas INFO Deleting /tmp/jblas949692298280708135/libjblas.so
– org.jblas INFO Deleting /tmp/jblas949692298280708135
No. Processing is going well.
Better to check, whether output files are created or not.
thank you for your answer @suribabu @ABraun
How can I get around the problem with orbit files in python splitting_slaves.py project.conf process?
I am getting this error when exporting stamp. How can I overcome?
…first_line_time metadata value is null
last_line_time metadata value is null
SNAP STDOUT:INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL 2.2.3 found on system. JNI driver will be used.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Installed GDAL 2.2.3 set to be used by SNAP.
Executing processing graph
INFO: org.hsqldb.persist.Logger: dataFileCache open start
…first_line_time metadata value is null
last_line_time metadata value is null
.10%…20%…31%…42%…52%…63%…74%…85%… done.
– org.jblas INFO Deleting /tmp/jblas2346160495396609768/libquadmath-0.so
– org.jblas INFO Deleting /tmp/jblas2346160495396609768/libjblas_arch_flavor.so
– org.jblas INFO Deleting /tmp/jblas2346160495396609768/libgfortran-4.so
– org.jblas INFO Deleting /tmp/jblas2346160495396609768/libjblas.so
– org.jblas INFO Deleting /tmp/jblas2346160495396609768
Had you checked the results?
The log says 85% …done.
So it seems it succeeded
thank you for your answer @suribabu
My last question: It covers all IW in the splitting slaves process, but can we split in 2 bursts ?