Please provide dates, orbit (precise or restituye) and AOI so I can try to replicate
I am afraid that could be beyond snap2stamps but exactly for that I need to check it first
Please provide dates, orbit (precise or restituye) and AOI so I can try to replicate
I am afraid that could be beyond snap2stamps but exactly for that I need to check it first
Hi @mdelgado i continue the test snap2stamps v2. I wonder How is efected master selection our tıme series results? And i made some examples. I prepared 105 S1A stacks for StaMPS and choose diffrent options for master selection like auto,manual and last. In a results for the same aoi i had 3 diffrent perpendicular baselines with the same results.
How should we interpret these results?
Note: To set the date in manual selection i used SNAP9.0 Insar Stack Overview.
Thanks…
Ty very much @mdelgado. The problem was caused by me and I solved it.
That indeed means that the master selection does not change the results on the underlying phenomenon that is being measured.
However, it interferes in more or less manner on the amount of PS points resulted.
This on the case of Sentinel data is less strong as it’s orbits are constrained and controlled and the Doppler difference is limited
Could you provide the PS numbers you got to illustrate that?
Thanks
In this study i used same threshold value for all stacks in StaMPS. (Da:0.20)
Master Selection Auto: 20200429
11225 PS points
Master Selection Manual: 20211009
11273 PS points
Master Selection Last: 20230309
11449 PS points.
Its looks like you are right!
But could you please give us some extra information about this explanation " This on the case of Sentinel data is less strong as it’s orbits are constrained and controlled and the Doppler difference is limited"
Thanks…
And when is tried this gpt -c 16G PROJECTFOLDER/graphs/coreg_ifg2run.xm this is what I get
Hi, @mdelgado!
It’s great to hear that there is a newer version of SNAP2StaMPS! Thank you and all the contributors for the effort.
Are you planning to create a video tutorial for it? Or is there already one available that I can access?
Cheers.
The error message says “Java heap space” so my guess is that increasing the value on the -c gpt option shall work.
Can you try it?
Honestly I have not thought to make a video tutorial, but if you believe it could help more people we may plan to have it.
In the meantime, which is the specific point you need to know more about?
I am sorry for my late reply. I believe a video tutorial could be extremely useful, especially for self-learners such as myself. It might also reduce the number of errors that users get caused by simple mistakes.
There is no specific point I wish to know more about, but I find video tutorials more helpful than user manuals in general, at least for beginners.
hello, I´m trying to run the “python topsar_step_2_splitting_secondaries.py -F project.conf” script but i got this error:
error: [NodeId: TOPSAR-Split] -1
I did the master topsar split at SNAP and located the product in the MasterSplit folder to continue with the secondaries topsar split but dont know how to solve this.
Please Help.
Hi Maria,
I strongly believe that the error is caused due to the fact that scihub has changed into Copernicus Data Space Ecosystem and the orbits can not be found.
However, if you update SNAP (the one installed with snap2stamps is under anaconda/envs/snap2stamps/snap/bin ) it shall be solved.
Until I will create a new conda package with SNAP updated each user shall do that manually.
Please try and let us know
Best
Hi, I did what you said but the it doesnt solved the problem. I modified the topsar_secondaries_split_applyorbit.xml file to set the subswath of interest area.
Hello.
now im trying to run the python topsar_step_3_coreg_ifg_topsar_smart.py -F project.conf step and again a have this problem.
please help. I cant run the earlier version because of python version …
I feel that the configuration of the project.config isnt clear and snap2stamps V2 Manual hasnt been updated, there are no tutorial videos, or any documentation to follow steps. these factors avoid more people to use this new version ![]()
Interesting! Thanks for your comments.
you should not modify the xmls by yourself. From the pic I see you changed the IW but you let the polarisation there.
from snap2stamps v1 I have seen that almost nobody read the Manual, so this is why it has not been updated. But you are right and I should work on it. However, if you read the README on github it says what you need to do to get it done. But as said… I will work in a simplify version of the Manual in the near future.
Thanks for your comments.
Hi,
I am trying to apply new version of snap2stamps on my dataset. However, I couldn’t manage the splitting process for both master and slaves. Here you can find the logs files and project_topsar.conf file. I am using Ubuntu 20.0 SNAP 9. Could you please tell me what I did wrong.
Thank you.
splitsecondaries.log (107.7 KB)
Hi!
Let’s try with this:
Please modify the MASTER line in the project file by removing everything after #
MASTER = /media/vboxuser/Elements/Test/master
instead of the
MASTER = /media/vboxuser/Elements/Test/master #example master D:/project/master/20220124.dim
I am also wondering whether your definition of the GRAPHFOLDER contains the graphs included within the snap2stamps package.
For the coregistration step, please ensure that the EXTDEM is properly defined as I see some Windows paths but the other ones are Linux paths.
As advice for you and others, ensure that the files you shared do not contain sensitive information, such as credentials etc ![]()
very excited about this!
I couldn’t quite tell from the discussion, but the Github details mention installation in Ubuntu 20.04 or 22.04, so does this new version not work on Ubuntu 18.04?
And for clarity, Python 3.11 is now required?
No, snap2stamps works on any OS (Linux, Windows, etc) but the installation dependencies I included with the sudo apt-get command will only work on Ubuntu 20, and 22, as for Ubuntu 18 they may have different names.
Regarding python, please use the conda environment file provided within the snap2stamps package, so all goes smoothly.
If you follow the installation instructions I hope you will find it easier than installing all the libraries from scratch, that was the idea.
Any recommendations if I’m getting the error discussed at the top of the thread but I am using Ubuntu 20.04?
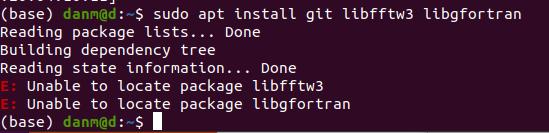
edit: seems I’ve successfully installed libfftw3 and libgfortran individually following the video linked above and the instructions on the Anaconda site. Could this result in any issues?
also, is there supposed to be a “.snap” folder in the /anaconda3/envs/snap2stamps directory? I only have the what I believe is the “snap” installation directory, though there is a “.snap” directory within the latter.
The auto_run script worked up until step_3…
Here is the output from the gpt command in case that’s useful
and my conf settings