Would it be possible to check that snap2stamps works with SNAP 10/11?
Dear @mdelgado ,
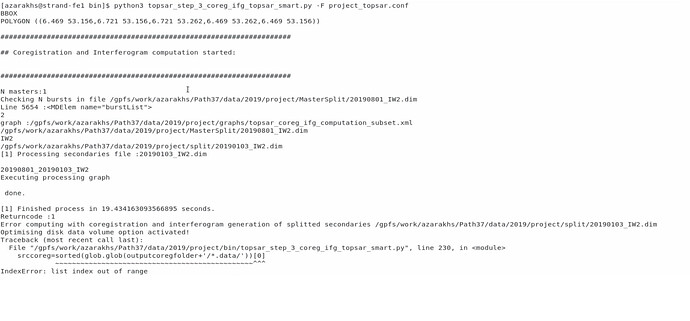
I am sorry to bother you. I followed your suggestion and kept EXTDEM empty. However, when I set SMARTHDD = Y, I encountered the following error during the coregistration and interferogram generation:
Error computing with coregistration and interferogram generation of splitted secondaries /gpfs/work/azarakhs/Path37/data/2019/project/split/20190103_IW2.dim
Optimising disk data volume option activated!
Traceback (most recent call last):
File “/gpfs/work/azarakhs/Path37/data/2019/project/bin/topsar_step_3_coreg_ifg_topsar_smart.py”, line 230, in
srccoreg=sorted(glob.glob(outputcoregfolder+‘/*.data/’))[0]
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~^^^
IndexError: list index out of range
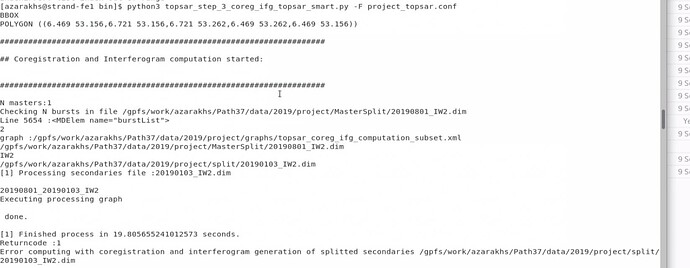
Additionally, when I set SMARTHDD = N, I received the following error:
Returncode :1
Error computing with coregistration and interferogram generation of splitted secondaries /gpfs/work/azarakhs/Path37/data/2019/project/split/20190103_IW2.dim
Hi everyone,
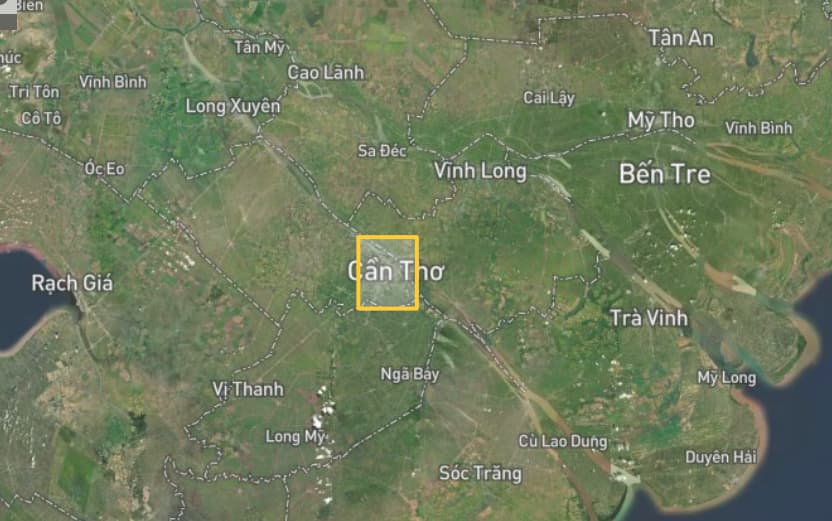
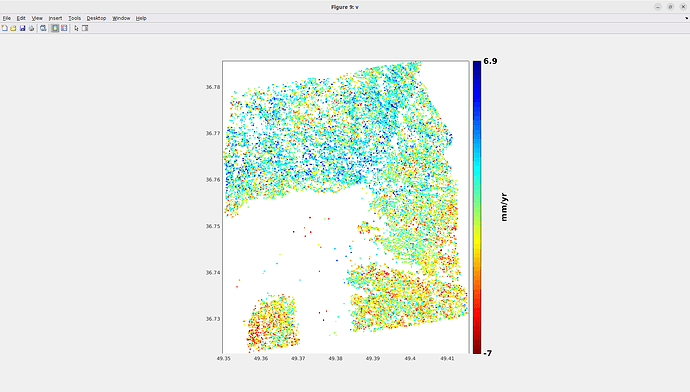
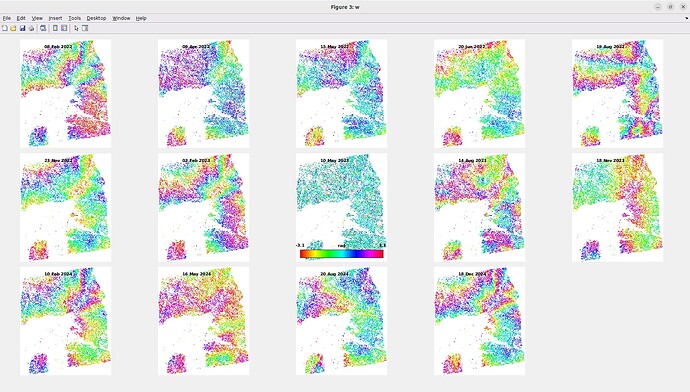
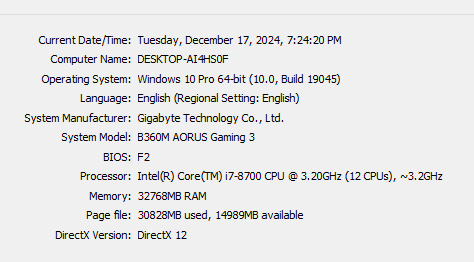
I encountered an issue at step_3 (I completed the previous steps without any problems). It is taking too much time to perform the coregistration between the Master image and the Slave images, even though my study area is not very large. Therefore, I am wondering whether the long processing time is due to a mistake I made in this step or simply because my computer is not powerful enough. I have attached my computer’s specifications, the completion time for each Slave image (~18 hours), the AOI (yellow rectangle). and my conf file.
Hi dear @mengdahl
Here is a work with SNAP 11 results, using ubuntu 22.
Thanks for your great job @mdelgado
In the old version of the Snap2stamps I used aps_linear command to compute the tropospheric delay map, but now when I use the command I face with this error :Error using load
Unable to read file ‘/home/…/INSAR_20230510/phuw.mat’. No such file or directory.
Error in aps_linear (line 92)
phuw = load(phuw_matfile);
Shouldn’t this file be created in the previous steps?
I guess that aps_linear shall be used after phase unwrapping (I suggest you check the StaMPS user manual for that).
I am not sure you run step 6 before trying aps_linear or not.
Yes, ypu are right. I did after but the same error
please check the existance of the file itself. If the file exists, I suggest you look into the code of aps_linear, maybe it searches in different path or so.
from other side, this thread is more focused on the snap2stamps package rather than StaMPS processes. So you may additional help in a dedicated thread for that.
I test it. It works both of them. But diffrent snap version subset diffrent areas. For the same aoi.
Hi @mdelgado
I wonder is it possible to process full IW or frame do not make subset in step5 with snap2stamps.
Best
Suat
Hi @suat,
It should be easy to implement it (next version of snap2stamps). Thanks for the feedback!
Today you could just remove the Subset operators from the topsar_export_mergeIW_subset.xml graph
Let me know if you need any help.
Best
Yes it is not be difficult. I changed a few line and its done.
Thanks.
Suat