Hello,
I’ve been working on a Sentinel 2 Image, resampling to 10m and getting spectral signatures, etc, and I’m having trouble with the Spectral Unmixing Tool under the Optical Tab. It seems it won’t allow me to write the endmember file anywhere on my computer.
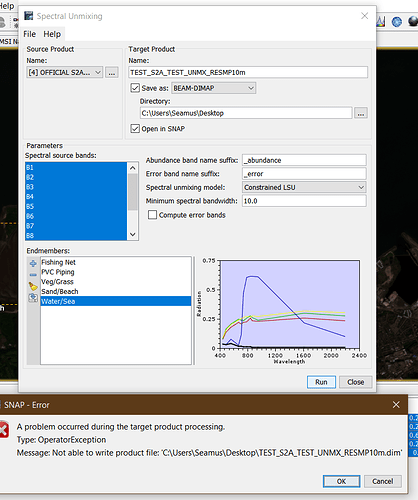
Here’s my settings and the initial error:
And here’s the code breakdown:
Error Details
java.io.FileNotFoundException: C:\Users\Seamus\Desktop\TEST_S2A_TEST_UNMX_RESMP10m.data\Veg\Grass_abundance.hdr (The system cannot find the path specified)
at java.io.FileOutputStream.open0(Native Method)
at java.io.FileOutputStream.open(FileOutputStream.java:270)
at java.io.FileOutputStream.<init>(FileOutputStream.java:213)
at java.io.FileOutputStream.<init>(FileOutputStream.java:162)
at java.io.FileWriter.<init>(FileWriter.java:90)
at org.esa.snap.core.dataio.dimap.EnviHeader.createPhysicalFile(EnviHeader.java:110)
at org.esa.snap.core.dataio.dimap.EnviHeader.createPhysicalFile(EnviHeader.java:92)
at org.esa.snap.core.dataio.dimap.DimapProductWriter.writeEnviHeader(DimapProductWriter.java:429)
at org.esa.snap.core.dataio.dimap.DimapProductWriter.getValidImageFile(DimapProductWriter.java:409)
at org.esa.snap.core.dataio.dimap.DimapProductWriter.createImageOutputStream(DimapProductWriter.java:443)
at org.esa.snap.core.dataio.dimap.DimapProductWriter.getOrCreateImageOutputStream(DimapProductWriter.java:392)
at org.esa.snap.core.dataio.dimap.DimapProductWriter.writeBandRasterData(DimapProductWriter.java:256)
at org.esa.snap.core.gpf.common.WriteOp.writeTileRow(WriteOp.java:430)
at org.esa.snap.core.gpf.common.WriteOp.computeTile(WriteOp.java:341)
Caused: org.esa.snap.core.gpf.OperatorException: Not able to write product file: 'C:\Users\Seamus\Desktop\TEST_S2A_TEST_UNMX_RESMP10m.dim'
at org.esa.snap.core.gpf.common.WriteOp.computeTile(WriteOp.java:375)
at org.esa.snap.core.gpf.internal.OperatorImage.computeRect(OperatorImage.java:82)
at javax.media.jai.SourcelessOpImage.computeTile(Unknown Source)Caused: org.esa.snap.core.gpf.OperatorException: Not able to write product file: 'C:\Users\Seamus\Desktop\TEST_S2A_TEST_UNMX_RESMP10m.dim'
at org.esa.snap.core.gpf.internal.OperatorExecutor$GPFImagingListener.errorOccurred(OperatorExecutor.java:381)
at com.sun.media.jai.util.SunTileScheduler.sendExceptionToListener(Unknown Source)
at com.sun.media.jai.util.SunTileScheduler.scheduleTile(Unknown Source)
at javax.media.jai.OpImage.getTile(Unknown Source)
at com.sun.media.jai.util.RequestJob.compute(Unknown Source)
[catch] at com.sun.media.jai.util.WorkerThread.run(Unknown Source)
The only extra information I can understand from this is that it’s saying the path/file doesn’t exist, but the Tool is supposed to create the file…?
This occurs even after my modest troubleshooting attempts:
- I thought maybe the file name was too long or had spaces (Windows has earlier had problems with the length of S2 filenames) so renaming to just “TEST_S2A_TEST_UNMX_RESMP10m” might have fixed it.
- I thought the initial default output directory (in “C:\Users\Seamus\.snap\auxdata\Sen2Cor-02.08.00-win64”) might have been write-protected somehow despite previous outputs being written there without issue, so I wanted to test somewhere with little write issues like the Desktop.
- I double checked I have ample space for writing the files on the hardrive (Over 500GB freespace)
Is there something obvious I’m missing to those with a better grasp of programming? It’s going to be something simple but I just can’t see it.
Thank you.