Hallo,
three months ago I used to run mt_prep_gamma, now I got errors.
I’m trying with mt_prep_snap but i get this one:
should I run first (again) a different StaMPS_CONFIG.bash?
thank you
Hallo,
three months ago I used to run mt_prep_gamma, now I got errors.
I’m trying with mt_prep_snap but i get this one:
should I run first (again) a different StaMPS_CONFIG.bash?
thank you
yes, every time you open the shell, you have to run StaMPS_CONFIG again (unless you include it in your bashrc, but this is not always a good idea)
I do not see the whitespace between the masterdate and the path to the INSAR folder. Please check.
Additionally, I am not sure, but it seems you are executing the mt_prep_snap in your current folder which is not the folder where stamps is, right? Please check
I hope this helps
Hi Abraun,
I completed my 8 steps of stamps processing. I felt some of my interferograms had snow cover and wanted to remove it from the processing. So I used ‘drop_ifg_index’ in the directory where all my patches are stored. I did not use this option for individual patches. Similarly I changed my ‘merge_resample_size’ to 10 in the directory containing all my patches.
Step 4 was successfully done but in step 5 PROCESSING PATCH_2 is the last message I see.
After this, there is no error nor it is moving forward to patch_3.
Following are my questions?
Regards
Narayanee
I have never used this option, but I guess it does not entirely remove the scenes from all steps, but rather exclude its contribution to the average velocity or the time series plots.
Maybe someone can comment here?
Referring to the variables: whatever parameter you change is valid for all patches. I recommend to stay in the top folder. StaMPS iterates through all patch folders automatically.
You can always check your current settings with getparms.
Yes, I did check using the getparm option. The value changes for the top folder but when I use getparm in individual patches, the value remains to be unchanged.
This is a major confusion now.
I have my following images after and before unwrapping.
Please do let me know about your insights about the quality of Interferograms and how to go about the interpretation of it.
With regards
Narayanee
Hi Abraun,
I changed my reference latitude and longitude in the stamps, but still the whole area displays.
The limits in the x and y axis change but the figure shows the complete area.
I have done all the 8 steps in stamps. But I still get the following error when I
type ps_plot(‘v-dao’)
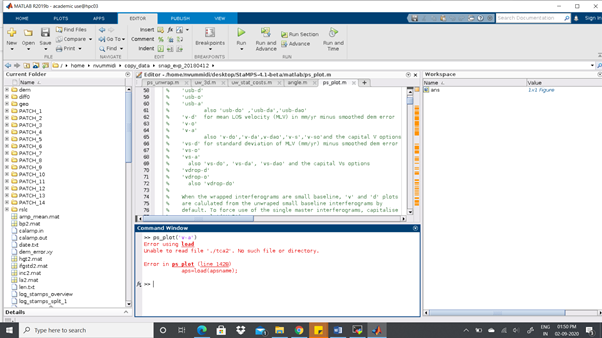
Can anyone please help me in this ?
did you set LON_RG and LAT_RG as part of the ps_plot command? What values did you give these variables?
You can see the documentation here: https://github.com/dbekaert/StaMPS/blob/master/matlab/ps_plot.m
(especially lines 108-110)
Hey Abraun,
I changed it using setparm option, but I did not change the values in the ps_plot.
I will try that. Thank you for your input.
It will be nice if you can give the input for the other question on ps_plot(‘v-dao’)
With regards
Narayanee
you can only use the a flag (in “v-toa”) if you have performed atmospheric correction using TRAIN or GACOS. If this is not the case, you can only plot “v-to”.
Thank you. I will look into it.
Same here, did you fix it?
I’m having the same problem, any luck?
Hi,
try to set setparm(“merge_resample_size”,0)
and run again stamps(5,5)
stamps(5,5) command didn’t executed due to following errors:
STAMPS: Will process patch subdirectories
STAMPS: ########################################
STAMPS: ################ Step 5 ################
STAMPS: ########################################
STAMPS: Directory is PATCH_1
PS_CORRECT_PHASE: Starting
Correcting phase for look angle error…
GETPARM: small_baseline_flag=‘n’
PS_CORRECT_PHASE: Finished
STAMPS: Directory is ulaanbaatar
PS_MERGE_PATCHES: Starting
Merging patches…
GETPARM: merge_resample_size=0
GETPARM: merge_standard_dev=Inf
Processing PATCH_1
Index exceeds matrix dimensions.
Error in llh2local (line 41)
dlambda=llh(1,z)-origin(1);
Error in ps_merge_patches (line 485)
xy=llh2local(lonlat’,ll0)*1000;
Error in stamps (line 474)
ps_merge_patches
Is there any solutions.?
ps_parms_initial.log (2.6 KB)
STAMPS.log (36.9 KB)
Try this below one.
solved by myself.
stamps should be run in PATCH directory. 
Not sure what you did… but if you run StaMPS within the INSAR_XXXXX folder, it takes cares all the subfolders and so on.
Probably you run step 5 or so within the PATCH folder, which lead to the issue you reported.
Happy to see that you solved it. This is the best part of it.
Enjoy StaMPS
hello I have the following error in step 8 of the processing with Stamps:
Index in position 1 exceeds array bounds (must not exceed 30350855).
Error in ps_scn_filt (line 179)
xy_near=ps.xy(ix1:ix2,:);Error in stamps (line 540)
ps_scn_filt
I use PAZ images and the following parameters:
(extremely educated guesses by BW and MS)
| Parameter | Default | Used |
|---|---|---|
max_topo_err |
20 | 10 |
filter_grid_size |
50 | 40 |
clap_win |
32 | 16 |
scla_deramp |
‘n’ | ‘y’ |
percent_rand |
20 | 1 |
unwrap_grid_size |
200 | 50 |
unwrap_time_win |
730 | 88 |
scn_time_win |
365 | 88 |
scn_wavelength |
100 | 50 |
unwrap_gold_n_win |
32 | 16 |
can someone help me to solve the error?
I have the same problem , tried with setparm(“merge_resample_size”,0) but still not working