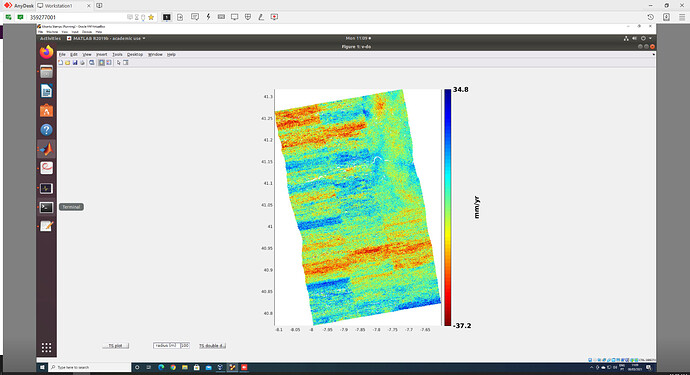
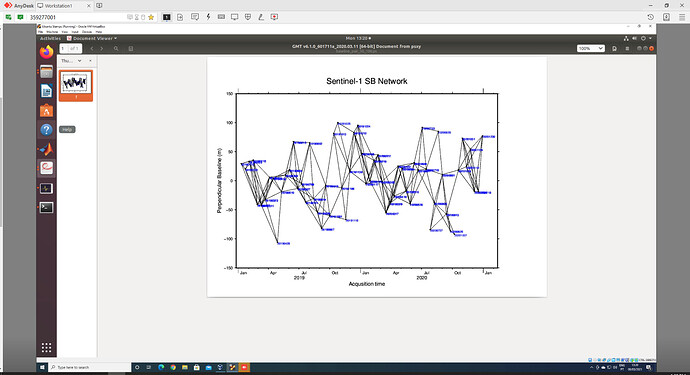
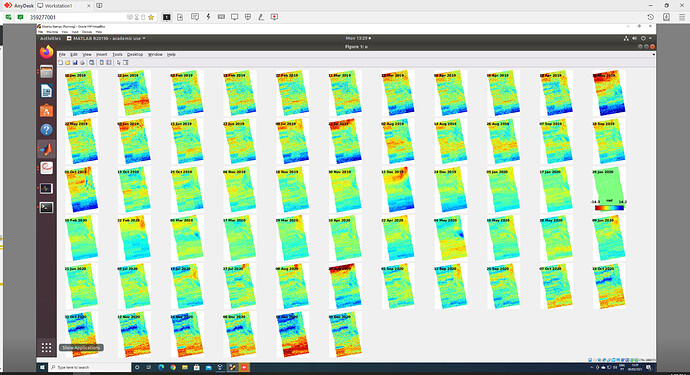
Hello all, I have just finished processing SBAS using GMTSAR + StaMPS and I get this very strange result:
ps_plot(‘u’) shows the same problem:
Does anyone recognize this anomaly?
Using the following params in Stamps with amplitude dispersion index of 0.6:
Created: '01-Mar-2021'
clap_alpha: 1
clap_beta: 0.3000
clap_low_pass_wavelength: 800
clap_win: 32
density_rand: 2
drop_ifg_index: []
filter_grid_size: 50
filter_weighting: 'P-square'
gamma_change_convergence: 0.0050
gamma_max_iterations: 3
gamma_stdev_reject: 0
heading: -13.5919
insar_processor: 'gmtsar'
lambda: 0.0555
lonlat_offset: [0 0]
max_topo_err: 20
merge_resample_size: 0
merge_standard_dev: Inf
n_cores: 8
percent_rand: 1
plot_color_scheme: 'inflation'
plot_dem_posting: 90
plot_pixels_scatterer: 3
plot_scatterer_size: 120
quick_est_gamma_flag: 'y'
ref_centre_lonlat: [0 0]
ref_lat: [-Inf Inf]
ref_lon: [-Inf Inf]
ref_radius: Inf
ref_velocity: 0
sb_scla_drop_index: []
scla_deramp: 'n'
scla_drop_index: []
scla_method: 'L2'
scn_deramp_ifg: []
scn_kriging_flag: 'n'
scn_time_win: 365
scn_wavelength: 100
select_method: 'DENSITY'
select_reest_gamma_flag: 'y'
shade_rel_angle: [90 45]
slc_osf: 1
small_baseline_flag: 'y'
subtr_tropo: 'n'
tropo_method: 'a_l'
unwrap_alpha: 8
unwrap_gold_alpha: 0.8000
unwrap_gold_n_win: 32
unwrap_grid_size: 200
unwrap_hold_good_values: 'n'
unwrap_la_error_flag: 'y'
unwrap_method: '3D_QUICK'
unwrap_patch_phase: 'n'
unwrap_prefilter_flag: 'y'
unwrap_spatial_cost_func_flag: 'n'
unwrap_time_win: 730
weed_max_noise: Inf
weed_neighbours: 'n'
weed_standard_dev: Inf
weed_time_win: 730
weed_zero_elevation: 'n'