I wish to apply a user defined image sharpening filter on a satellite image but I cannot find Image Filter option in Raster menu even when the image is open in View.
I have created a 5x5 matrix as required.
Please help
You can’t see Filtered Band in the Raster menu?
And what happens if you enter “Filter” in the search box at the upper right?
And how have you created the matrix if the menu item is not there.
Maybe your installation is somehow broken?
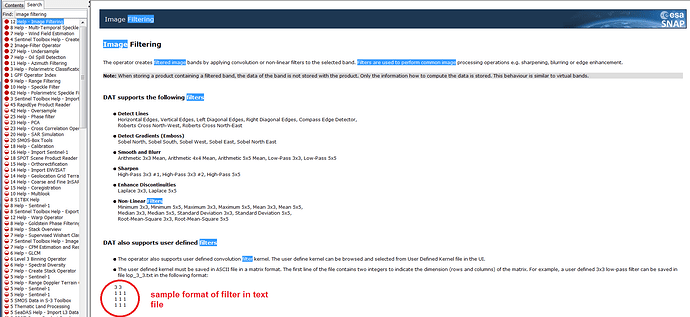
Dear Marco,Thanks for your response.I think that I am lost with the terminology being used?The attached snapshot of Help shows format of a 3x3 matrix which can be used as a filter over an image. On the same ground I created a text file of 5x5 matrix. As the attached Help snapshot shows, I expect Image Filtering option under Raster, which is missing in my case. I assume that Image Filter option will allow me to choose the 5x5 matrix file which I created.I hope I am able to get through my problem to you.Look forward to your guidence on this.Thanks once again for your help. Yogdeep Desai
Yes it seems that Image Filtering got lost. The documentation is also pretty old. It still mentions DAT. Maybe @lveci can say where this option is gone.
But you can do the same with the Filtered Band function. You also have a graphical user interface for defining the kernel.
If you want to use the already stored file you can use Image-Filter from the command line.
Type gpt Image Filter -h for more information.
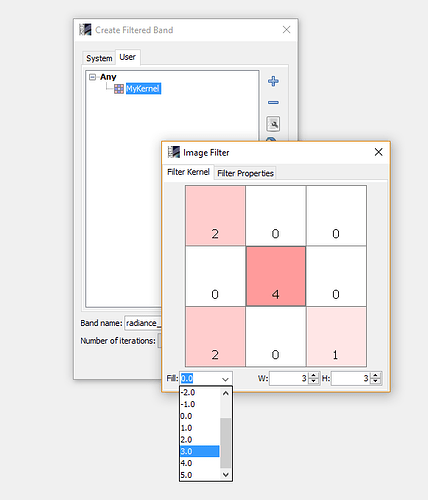
Dear Marco,That was pretty quick response. In the beginning, even I was hopeful of able to create filtered bands from the Filtered Band >User option. Unfortunately the kernel window which you have shown do not pop-up at all (in fact nothing happens on clicking MyKernel).I might have to look for fresh installation or, may be, a latest version.Thanks once again for your support.Regards,
Yogdeep Desai
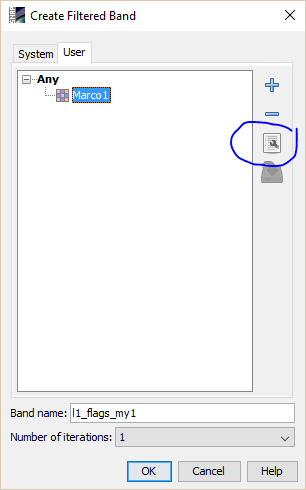
You need click on the marked button.
Sorry to dig this out.
Can I apply gpt Image-Filter with more than 7x7 kernels and 5 iterations
The menu only allows 5 iterations:
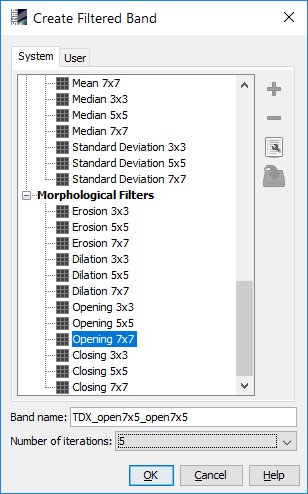
How would the gpt command look like?
Modules presented in SNAP deskstop are not a perfect replica of what you can do in gpt. Or at least according to their description.
I don’t think you can do more than one iteration in gpt. If you write your commands in .sh file, just loop over a temporary file
To use 7x5 opening filtering, you have to create your own filter. You can specify the size and your operation (in your case, OPEN).
gpt Image-Filter -h suggests that you can use your newly created filter : (userDefinedKernelFile)
Usage:
gpt Image-Filter [options]
Description:
Common Image Processing Filters
Source Options:
-SsourceProduct=<file> Sets source 'sourceProduct' to <filepath>.
This is a mandatory source.
Parameter Options:
-PselectedFilterName=<string> Sets parameter 'selectedFilterName' to <string>.
-PsourceBands=<string,string,string,...> The list of source bands.
-PuserDefinedKernelFile=<file> The kernel file
Graph XML Format:
<graph id="someGraphId">
<version>1.0</version>
<node id="someNodeId">
<operator>Image-Filter</operator>
<sources>
<sourceProduct>${sourceProduct}</sourceProduct>
</sources>
<parameters>
<sourceBands>string,string,string,...</sourceBands>
<selectedFilterName>string</selectedFilterName>
<userDefinedKernelFile>file</userDefinedKernelFile>
</parameters>
</node>
</graph>
However, when I use the command
gpt Image-Filter ‘/home/qg/Documents/MIMO/coh tracking small test/Stack.dim’ -PselectedFilterName=“qg1” -PuserDefinedKernelFile=’/home/qg/.snap/auxdata/image_filters/user-filters.xml’ -t "/Documents/MIMO/test"
I’ve got this error :
Error: Incorrect file format: user-filters.xml
thank you for your comment!
I also was at the point to enter the graph manually but had similar issues with the calling of the filter.
@marpet already helped me in the past with non-documented gpt functionalities. Maybe he can help us  it should be possible, since there’s a parameter related to the use of user-defined kernels. Just which file are we supposed to give if it’s not the xml one.
it should be possible, since there’s a parameter related to the use of user-defined kernels. Just which file are we supposed to give if it’s not the xml one.
Indeed the Image-Filter operator is not well documented and it does not have the same functionality as the GUI “Create Filtered Band”.
We have it on our agenda to harmonise all these inconsistencies, but this will take a while.
The non-linear and morphological filters can’t be used from the command line. That’s not implemented.
Regarding the file format:
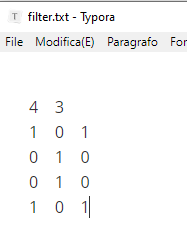
It must be like the attached file:filter.txt (31 Bytes)
On the first line the number of rows and columns. Following lines the values. All tab separated, or on separate lines.
The number of iterations is fixed to one. Probably you can overcome this limitation by using multiple Image-Filter operators in a chain.
I once wrote an example script which shows how filtered bands can be generated with Python. Maybe it is useful.
create_filtered_band.py (2.0 KB)
thank you!
Dear Mr. @marpet,
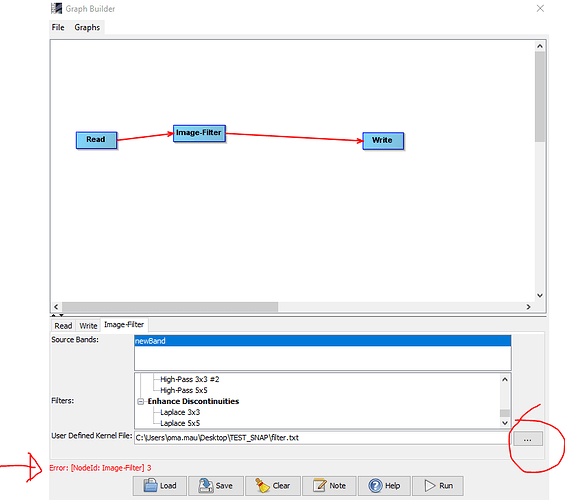
regarding the possibility to use a User Defined Kernel File (I mean, an user-defined filter…i’m interessing on Opening filter in particular) on the GraphBuilder. Could you please provide any feedback/commenst on the following steps that i’m performing (but unsuccessfully):
- create preliminarily the filter function required (accordingly with the rules described from you in this post) and save it as .txt file;
- From the GraphBuilder function named “Image-Filter”, skip the section of default filters and call the .txt file clicking the option User Defined Kernel Filter;
Could anybody confirm that a User Defined Kernel File should be a customized Filter in .txt format?
In the GraphBuilder and on the command line only linear filters are implemented. Opening belongs to the Non-linear filters and is not implemented.
Yes, the txt file is correct. Two things you can try.
Check if the tab separator wasn’t replaced by spaces by your editor and maybe add a linebreak at the end of last row.
Dear Mr. @marpet,
thanks for your answer. Starting from the point that Non-linear filters are not implemented on GraphBuilder (indeed they are not available on the drop-down menu of Image-Filter function), I’d like to better understand the feasibility of a workaround. I’ve two questions:
-
Is it possible to write a Non-linear Filter (Opening in particular) following the rules described from you here Where is Image Filter option in Raster menu? ?
-
[In case of point one is feasable] Would be possible to run successfully the Opening filter in .txt format by the GraphBuilder?
Many thanks
Has this issue not been solved yet? I just want to create a min and max 3x3 filter. I can’t possibly go through 108 files and apply the filter separately?
Python’s pillow library provides a rank filter. Using ESA SNAP’s python interface, snappy, you should be able to create a python script to load a product, apply the pillow rank filter, and save the resulting filtered product. You can then use a batch script to run your python script on a list of files.
As i can’t find documentations on snappy and data conversion from org.esa.snap.core.datamodel.Band to Image so that PIL.ImageFilter can be applied, is there any possibility to show a small code example on how to convert the Band to the proper type and convert back after using a filter on it?
There are examples in this forum (e.g., using numpy with ESA SNAP snappy), and many examples using numpy arrays with PIL elsewhere.
Thank you for this script! I do have one question, though. How should I edit that to receive a 3x3 mean filter? I have basically no experience with that, so I don’t know how to even start, and as far as I have researched, snappy still doesn’t support non-linear filters.