thank you, these are great news. I would be very interested in this preliminary version, too!
see: https://bluestacks.vip/ , https://kodi.software/
To be extra clear:
i have not yet found any succes of using StaMPS on Windows by compiling it, i would highly recommend to skip the painful trial and error and move to linux. can ofcourse be dual boot as well.
Yes, it seems windows has not only problem in this terms, however, python and related issues, Linux is the solution.
Dear all.
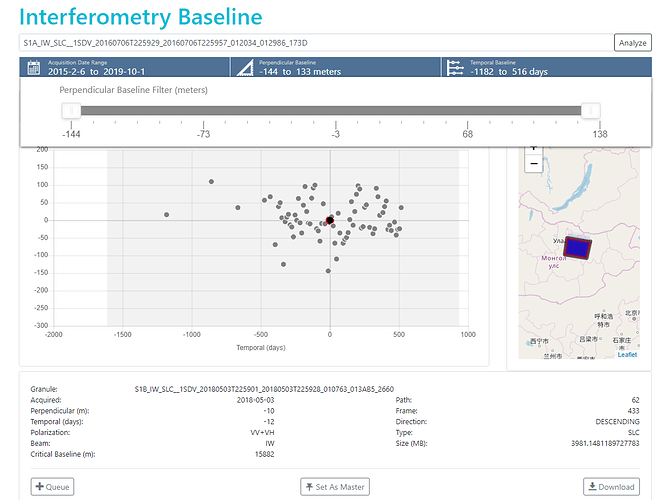
According to many guidelines, i have to choose optimal master image near time center and average baseline from collected data. So how about my choice showing on this figure?
for single-master approaches, one which lies in the temporal and perpendicular center is mostly selected. I would probably have taken the same.
Anyone faced into this?
SNAP STDOUT:b"INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters\nSEVERE: org.esa.s2tbx.dataio.gdal.activator.GDALDistributionInstaller: The environment variable LD_LIBRARY_PATH is not set. It must contain the current folder ‘.’.\nINFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.\nExecuting processing graph\nINFO: org.hsqldb.persist.Logger: dataFileCache open start\n done.\n\nError: [NodeId: Enhanced-Spectral-Diversity] Operator ‘SpectralDiversityOp’: Unknown element ‘useSuppliedShifts’\n-- org.jblas INFO Deleting /tmp/jblas1601710227008905183/libjblas.so\n-- org.jblas INFO Deleting /tmp/jblas1601710227008905183/libjblas_arch_flavor.so\n-- org.jblas INFO Deleting /tmp/jblas1601710227008905183\n"
Probably you are working with a single burst, right? If so, delete the ESD operator from the xml
I believe your error will disappear
Great!!!. Thank you
It works, after edited 2 xml file like this.
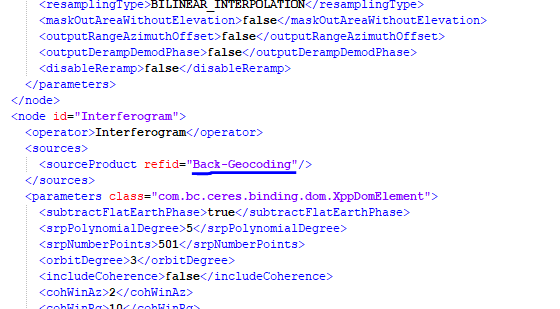
But is there any difference side of processing result quality?
No difference.
If you work only with 1 burst, ESD must not to be applied.
hallo I follow your workflow to prepare sentinel images for stamps processing.
Everythings seems ok but no insar_master_date folder was created!
The mt_prep_snap doesn’t find the slc.par files.
do you know wherw I was wrong?
I’m sorry, maybe is a simple thing but I’m new in this field! 
thank you so much
F.
just to go sure: did you run stamps_export.py as the last script?
also to go sure: the folder name will not contain “master_date” but the date of your master in this order: yyyymmdd. For example, the folder will be INSAR_20190721 for a master image selected on 21.07.2019.
I followed https://gitlab.com/Rexthor/gis-blog/blob/master/StaMPS/2-3_snap2stamps.md those steps.
And final export result was like this
I have used:
- 53 images.
- Stamps v4.1b, Snap 7, ubuntu server 16.04, gcc 4.8, matlab 2016b, python 2.7
- IW2, single burst
- All previous steps didn’t get any errors
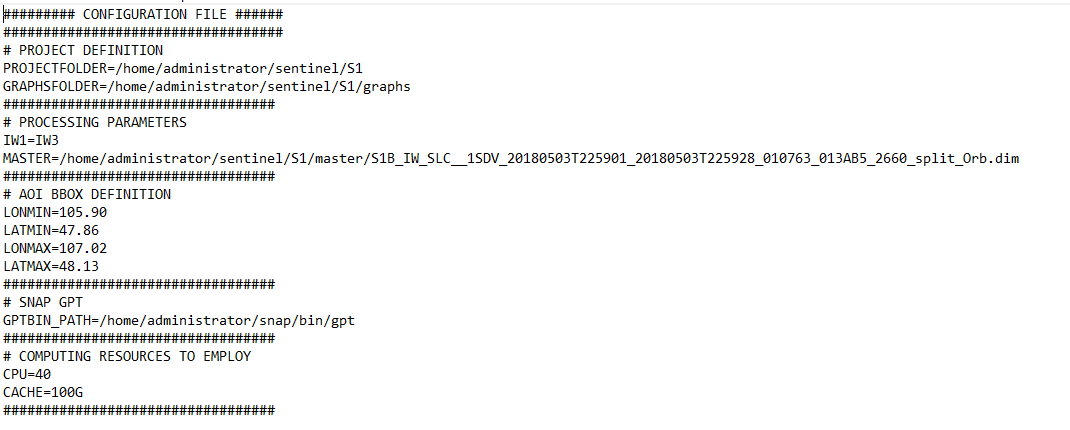
- Here is my config file.
- mt_prep_snap script doesn’t work. My results empty in my patch_1 folder
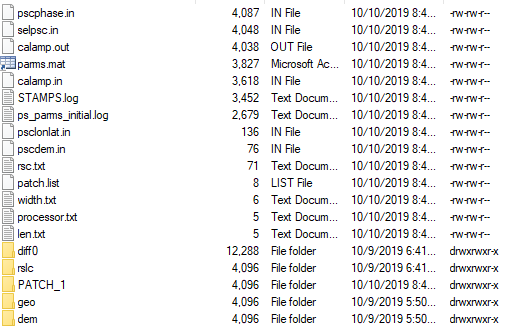
can you please also share the mt_prep_snap command you used?
Regardless of the error, I recommend creating a separate folder for the StaMPS processes. Simply create a new directory (at any location) and execute mt_prep_snap here. This helps you to keep the structure clean and doesn’t mix up the original files exported from SNAP with the ones created by Matlab.
Please share also the calamp.out … at it shows if any slave contains all 0’s.
Hi all
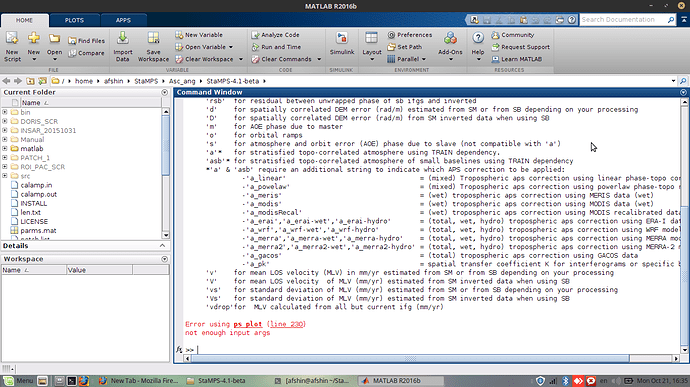
I have a problem with ps_plot and I face with this error
does ps_plot.m file have problem?
how can I fix it ?
even after step 6 when I want to plot ps_plot (‘v-dm’), this error occur.
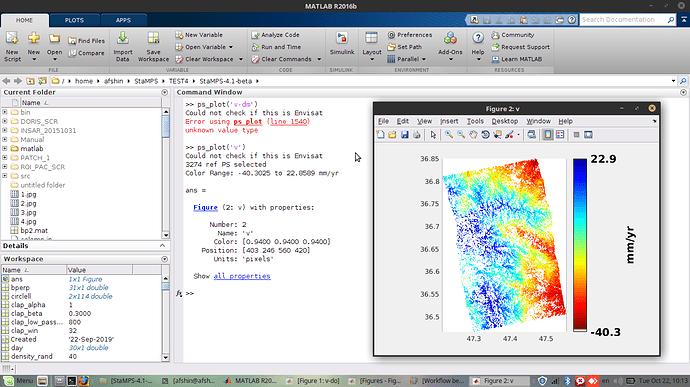
try ps_plot(‘v-dm’,1)
I tried but still doesn’t work
can you please post the command you entered in here? Maybe you use the wrong quotation marks or have a space where there should be none.