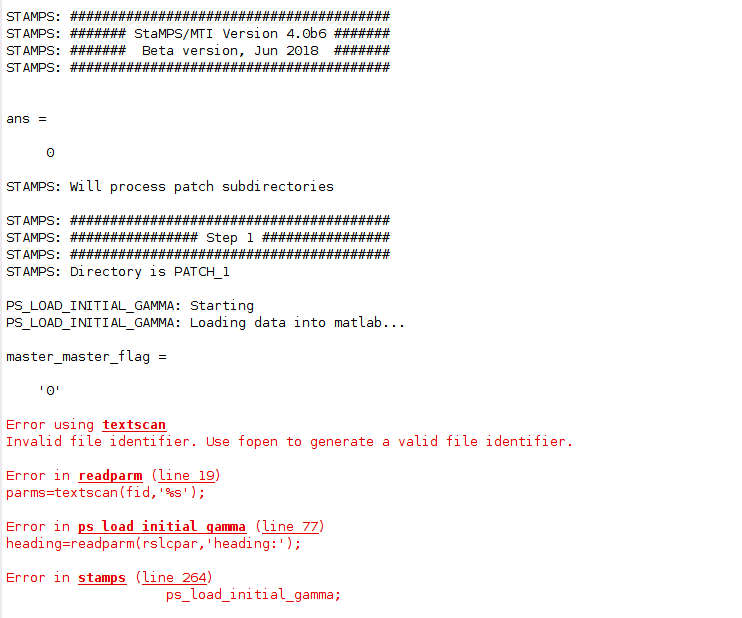
when i use viStamps to disaplay the result(linux),the unwrap phaset shows:
!(file:///C:\Users\leslie\Documents\Tencent Files\951755015\Image\C2C\NMUMR4ZSNB[%P[[X%`@Y@R.jpg)
D4P%6O0KG8Y)MOU@E4.jpg)
but the deformation shows:

F0N[I_M0Q)8R.jpg)
my deformation map is a strip,and be differerce from the manual of vistamps, and dont have a map layer
Sorry but I have never used viStaMPS.
Maybe anybody can give any hint?
My only guess it that you had not provided latitude and longitude information.
thank you,and i have try to vilisualizer the result by Stamps Visualizer (successfully) and iVstamps (failed),when i export the all the points to csv ,then load csv by R studio,it is really bad the page refresh very slowly because of the data volume,i wander how i can i control the step when export to csv file like export to a kml file
Well… not sure but indeed viStaMPS has nothing to do with the workflow between SNAP and StaMPS…
So I would suggest the proper forum fhreads that address the different issues.
Good luck with viStaMPS.
Still StaMPS has own commands to convert output to kml files: ps_gescatter
thanks for your reply
Dear mdelgado
i meet a mistake when i creat stack in snap:
after creat a stack of 22 S1 data, i check each intensity image in the stack and found that one(20150924) of the intesity image shows black ,what is happened to this situation?and when i do apply orbit file , all data 's orbit can be found in C:\Users\leslie.snap\auxdata\Orbits directory except the 20150924.so i think the black image was caused by failed to apply the orbit file in this data(20150924),but all of these steps in snap did not have a error message,what should i do?
Dear @Gijs,
Did you find a solution to solve this problem?
Hey Jennifer,
The problem seemed to be caused by an incorrect ifg, when i reprocessed the issue was solved. you can check this by checking the mean amplitude and ammount of 0s printed out at the mt_prep_snap phase.
In my case i had accidentally left an ascending aquisition in my descending folder or something like that.
Greetings,
Gijs
Thank you…will check such in my case…
What you mean by this? Whal should i check in mean amplitude and the amount of zeroes? I get some values… but what you mean by checking it? Can u tell me precisely please…
i get this error…
Could anybody tell me a feasible solution?
@Gijs just now i figured that one of my interferogram is blank with no values and all zeroes… Thank you
This is exactly what i said in the post above.
You had to check the print output given when you run mt_prep_snap.
but you posted the print ouput from StaMPS (1,1) print output.
Glad you found the problem either way, i was on vacation.
Greetings,
Gijs
Dear @andretheronsa @ABraun,
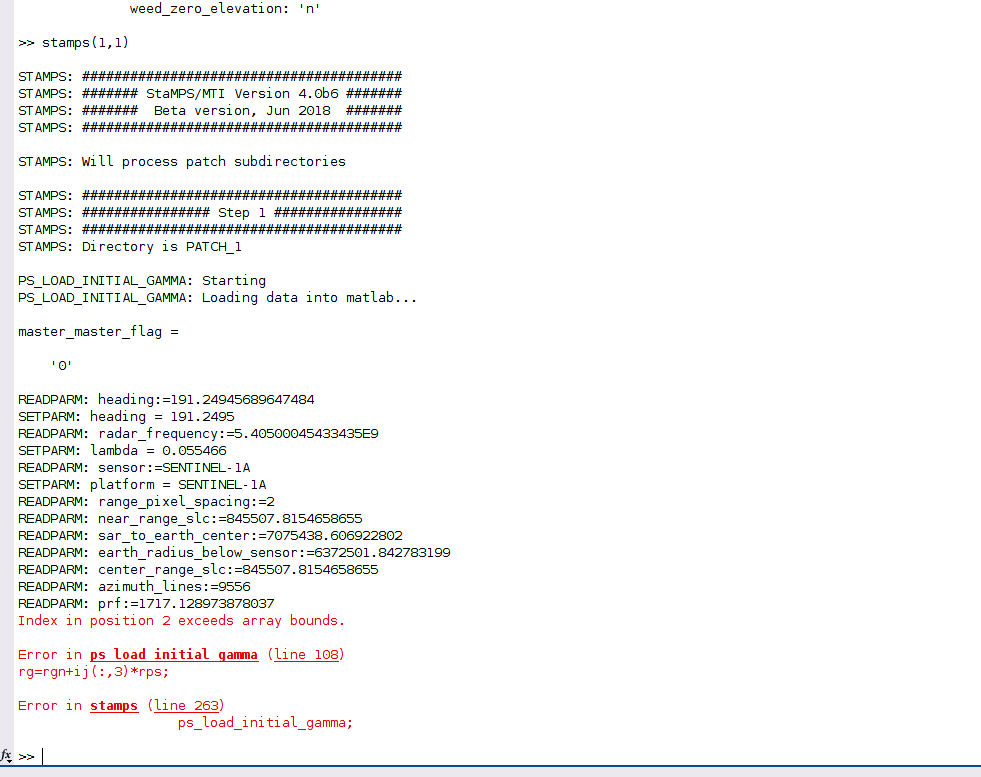
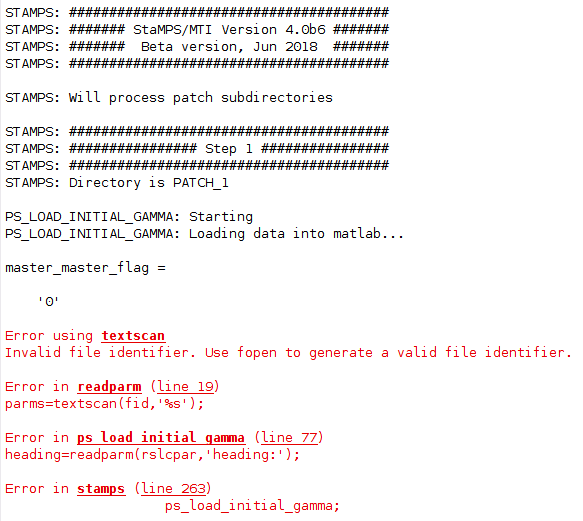
I have the same problem as you described here, in step 1 of StaMPS loading data into Matlab:
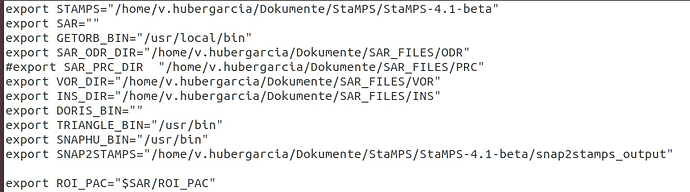
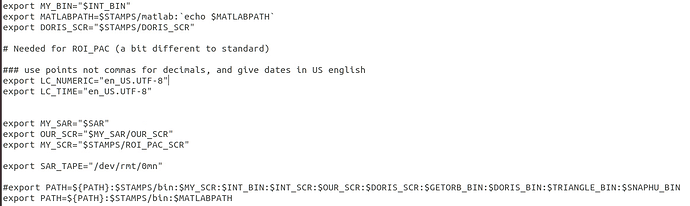
Did you find a solution for it? Could it be related to my StaMPS_CONFIG.bash, which I am not sure if I parametrised correctly?
I changed the last line according to the explanations given by @thho but I am not sure if this is the case for me.
I source StaMPS_CONFIG.bash, navigate to the folder containing the StaMPS input in the terminal and than start matlab. getparm() also works.
I am using:
- Ubuntu 18.04 LTS
- StaMPS 4.1 beta
- Matlab R2019a update 3
I would appreciate your helpt very much!
I can’t remember how I got rid of this error, but I remember that it was not the CONFIG file but rather related to the stamps scripts.
Did you use the distribution from here: https://github.com/dbekaert/StaMPS
Hello @ABraun,
yes I am 90% sure I did. Could it help to reinstall it?
Thanks.
it is worth a try.
From your screenshot I see that you haven’t defined a triangle and snaphu directory. If the executables are in the bin folder, everything is fine. I just wanted to go sure because this would cause problems in later steps of StaMPS.
Are you running on a windows machine by chance? I remember that I had this on a windows computer only.
Hi. No, I am using linux. I tried to reinstall StaMPS (unzipping the folder, navigate to the StaMPS/src folder, make, make install. Do I miss something?) but the error is still occuring.
Any further ideas? Thank you for your help.
if I recall correctly, the problem was already caused in the mt_prep_snap part before matlab was started.
But I have no idea how to check if the interferogram are generated correctly there.
Did you repeat this step as well after reinstalling StaMPS? (removing the old folder before, of course)