Hi, I’m new in snap. I want to do a supervised classification with Random Forest Classifier but when I execute the tool I receive the notification “bound must be positive”. How can I solve this?
reproject your data to WGS84 (uncheck “reproject tie-point grids”) and try again. Somehow, the classification module cannot handle UTM coordinates.
See here: Rndom forest classification steps
It worked, thank you very much!.
Kind regards
Hi, I am working with SNAP to classify crops. I got same error(“bound must be positive”).
And I tried to do what ABraun said as rectification(uncheck “reproject tie-point rids”). but I am unable to find this option can you please help me in this ?
Raster > Geometric > Reprojection
Select Geographical (lat/lon) WGS84 here.
Hi @ABraun and @Misio, Recently I am following the solution from this discussion, but I found the same error for both GRD and SLC Sentinel 1 and Sentinel 2 after reprojecting to WGS 84 while doing the random forest classification. I also found the same error while using similar data in UTM WGS 84 zone 50S. How to fix this issue?, Thank you.
Sentinel-2 data is stored in UTM by default. So it is advisable to reproject it to WGS84 before merging it with Sentinel-1 (also WGS84 after terrain correction). Have you seen this tutorial? Synergetic use of S1 (SAR) and S2 (optical) data and use of analysis tools
Profesor @ABraun, thank you for your reply. It is very important for my study. I forgot to explain in details that I did random forest for S1 and S2 in two different processes, not merging both data for this time. But, I will check the suggested tutorial soon. Thank you
could it be that the training data (maybe imported as SHP) are stored in UTM coordinates?
I used the training samples that previously made in QGIS as shapefile and it stored in WGS84 .
after which step did you import the training samples?
Professor @ABraun I have done all the pre-processing steps in order to run a Random Forest Classification.
My pre-processing steps are:
- Resampling (S2_Resampling) (Whole Image)
- Reprojection to WGS84 (Whole Image)
- Import vector and mask off the AOI
- Import Vector and mask off (artificial structures)
- Import Vector (Training samples)
- Random Forest Classification (5000 training samples) (10 number of trees)
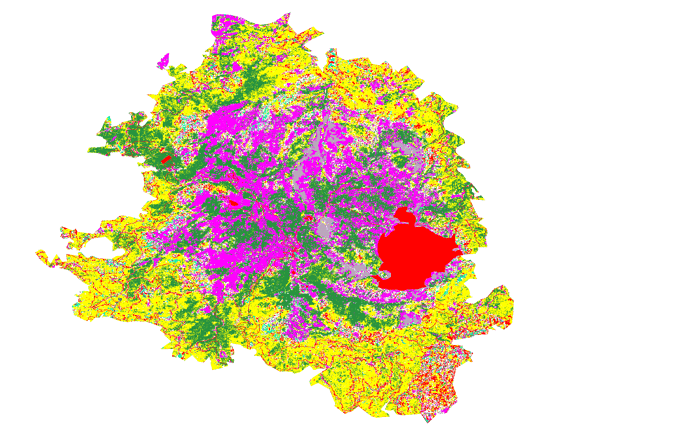
The result of this procedure is that, the system gives color (red) outside of my AOI and I solve this by masking again the AOI but, there are empty pixels which are not supposed to be empty but they have to show a value.
I have delete all the training samples which are overlayed and outside of the AOI lest they affect negatively the result.
I do not know what else I am supposed to do. Thank you in advance and for your patience.
The red color in the image it’s supposed to be NaN values as they are artificial structures.
PS. I have 7 classes to classify thus; maybe I have wrong to the number of trees and samples decision?
- Number of mask pixels : 1081315
-
Mask area: 90.207 km2
Full image
Example of empty pixels (White pixels)
please have a look at the remarks on unclassified pixels after Random Forest on page 24 of this tutorial: Landcover classification with Sentinel-1 GRD
The shapefile (training sample) imported after the polarization decomposition and backscatter (dB) processes completed.