Dear All,
I’m sorry for the lots of questions in this some time but I’m becoming a bit desperate.
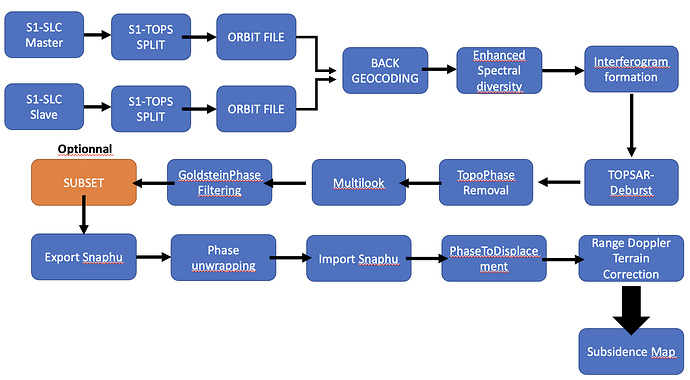
My goal is to doing this stuff with Snappy :
So I just wanted to separated the filter part and the interferogram formation just to control.
Here my code :
import sys
import os
from snappy import GPF
from snappy import ProductIO
from snappy import HashMap
from snappy import jpy
from snappy import WKTReader
import subprocess
import glob
import shutil
import gc
import time
import multiprocessing
import logging
def read_product(*args, **kwargs):
“”“Open product.
Arguments:
Accepts: .zip, .dim, unzipped .SAFE directory
*args
object from class fetchme.Product() with attribute ‘path_and_file’
**kwargs:
path_and_file
Returns:
product
Examples:
# EX1: case where argument a keyword argument
import snapme as gpt
p = gpt.read_product(path_and_file = ‘/path/file.zip’)
# EX2: case where argument is an object with attribute ‘path_and_file’
import fetchme
import snapme as gpt
obj = fetchme.Product()
p = gpt.read_product(obj)
# See native funtions available:
p.
EX: p.getName()
“””
#logging.info(‘reading product’)
# case if args agument passed
if len(args) > 0 and hasattr(args[0], 'path_and_file'):
pnf = args[0].path_and_file
# case if keyword agument passed
if len(kwargs) > 0 and 'path_and_file' in kwargs:
pnf = kwargs['path_and_file']
product = ProductIO.readProduct(pnf)
return product
def read(filename):
return ProductIO.readProduct(filename)
def get_name(obj):
r = obj.getName()
return
def write_product(obj, f_out=None, p_out=None, fmt_out=None):
if f_out is None:
f_out = get_name(obj)
if p_out is None:
p_out = '../data/'
else:
p_out = os.path.join(p_out, '') # add trailing slash if missing (os independent)
if fmt_out is None:
ext = '.dim'
fmt_out = 'BEAM-DIMAP'
else:
ext = ''
logging.info('saving product')
# print(p_out + f_out + ext)
# print(fmt_out)
# return
ProductIO.writeProduct(obj, p_out + f_out + ext, fmt_out)
#os.chdir(r’/Users/clement.goldmann/SAR_docker’)
def write_product2(obj, filename):
ProductIO.writeProduct(obj, filename , “BEAM-DIMAP”)
def topsar_split(product, IW):
# Hashmap is used to provide access to all JAVA operators
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(‘subswath’, ‘IW2’)
parameters.put(‘selectedPolarisations’, ‘VV’)
parameters.put(‘firstBurstIndex’, 1)
parameters.put(‘lastBurstIndex’, 3)
return GPF.createProduct(“TOPSAR-Split”, parameters, product)
def apply_orbit_file(product):
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(“Orbit State Vectors”, “Sentinel Precise (Auto Download)”)
parameters.put(“Polynomial Degree”, 3)
parameters.put(“continueOnFail”, True)
return GPF.createProduct(“Apply-Orbit-File”, parameters, product)
def back_geocoding(product):
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(“Digital Elevation Model”, “SRTM 3Sec (Auto Download)”)
parameters.put(“DEM Resampling Method”, “BICUBIC_INTERPOLATION”)
parameters.put(“Resampling Type”, “BISINC_5_POINT_INTERPOLATION”)
parameters.put(“Mask out areas with no elevation”, True)
parameters.put(“Output Deramp and Demod Phase”, True)
return GPF.createProduct(“Back-Geocoding”, parameters, product)
def EnhancedSpectralDiversity(product):
parameters = HashMap()
parameters.put(“Registration Window Width”, 512)
parameters.put(“Registration Window Height”, 512)
parameters.put(“Search Window Accuracy in Azimuth Direction”, 16)
parameters.put(“Search Window Accuracy in Range Direction”, 16)
parameters.put(“Window oversampling factor”, 128)
parameters.put(“Cross-Correlation Threshold”, 0.1)
parameters.put(“cohThreshold”, 0.3)
parameters.put(“numBlocksPerOverlap”, 10)
parameters.put(“esdEstimator”, ‘Periodogram’)
parameters.put(“weightFunc”, ‘Inv Quadratic’)
parameters.put(“temporalBaselineType”, ‘Number of images’)
parameters.put(“maxTemporalBaseline”, 4)
parameters.put(“integrationMethod”, ‘L1 and L2’)
parameters.put(“doNotWriteTargetBands”, False)
parameters.put(“useSuppliedRangeShift”, False)
parameters.put(“overallRangeShift”, 0.0)
parameters.put(“useSuppliedAzimuthShift”,False)
parameters.put(“overallAzimuthShift”,0.0)
return GPF.createProduct(‘Enhanced-Spectral-Diversity’, parameters, product)
def interferogram(product):
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(“Subtract flat-earth phase”, True)
parameters.put(“Degree of "Flat Earth" polynomial”, 5)
parameters.put(“Number of "Flat Earth" estimation points”, 501)
parameters.put(“Orbit interpolation degree”, 3)
parameters.put(“Include coherence estimation”, True)
parameters.put(“Square Pixel”, True)
parameters.put(“Independent Window Sizes”, False)
parameters.put(“Coherence Azimuth Window Size”, 2)
parameters.put(“Coherence Range Window Size”, 10)
return GPF.createProduct(“Interferogram”, parameters, product)
def topsar_deburst(product):
HashMap = jpy.get_type(‘java.util.HashMap’)
parameters = HashMap()
parameters.put(“Polarisations”, “VV”)
return GPF.createProduct(“TOPSAR-Deburst”, parameters, product)
def FirststepSAR(filename_1, filename_2):
print('Reading SAR data')
product_1 = read(filename_1)
product_2 = read(filename_2)
print('TOPSAR-Split')
product_TOPSAR_1 = topsar_split(product_1, 'IW2')
product_TOPSAR_2 = topsar_split(product_2, 'IW2')
print('Applying precise orbit files')
product_orbitFile_1 = apply_orbit_file(product_TOPSAR_1)
product_orbitFile_2 = apply_orbit_file(product_TOPSAR_2)
print('back geocoding')
backGeocoding = back_geocoding([product_orbitFile_1, product_orbitFile_2])
print('ESD')
ESD = EnhancedSpectralDiversity(backGeocoding)
print('interferogram generation')
interferogram_formation = interferogram(backGeocoding)
print('TOPSAR_deburst')
TOPSAR_deburst = topsar_deburst(interferogram_formation)
write_product2(TOPSAR_deburst, 'S1A_IW2_SLC__20230122_20230215_Orb_Stack.dim')
print('Operation successfull')
and I run this one :
from DefineSar import FirststepSAR
FirststepSAR(“S1A_IW_SLC__1SDV_20230122T111531_20230122T111559_046895_059FB4_89F7.zip”,“S1A_IW_SLC__1SDV_20230215T111530_20230215T111558_047245_05AB64_F1B0.zip”)
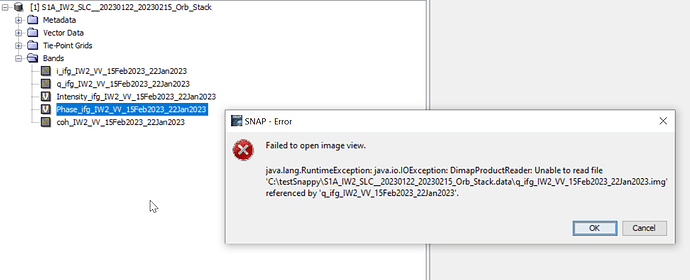
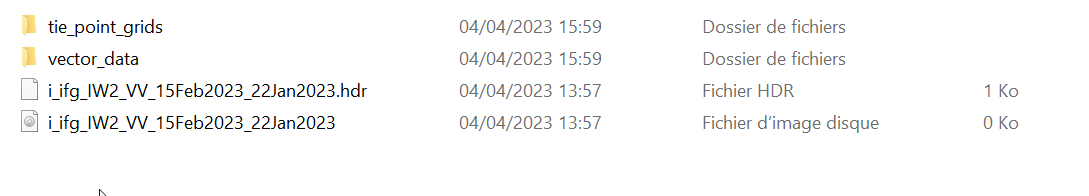
The trouble is when I wanted to open in snap on my other computer I’m unable to read it and I have this trouble : Failed to open image view
java.lang.RuntimeException DimapProducReader : Unable to read file
Maybe snappy doesn’t allow us to see the result directly in SNAP and I just product the result instantly. So I need the data folder or maybe it’s just impossible.
Cheers,
Clément