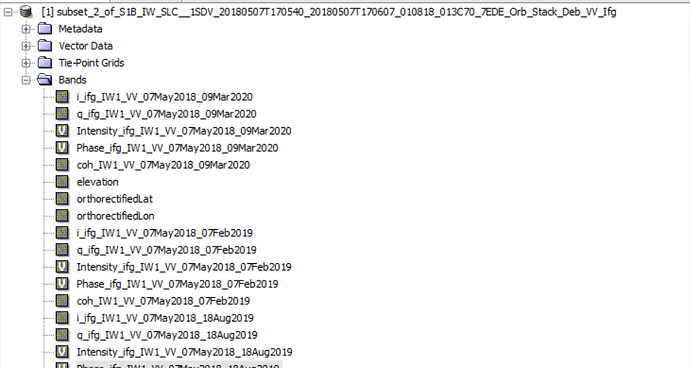
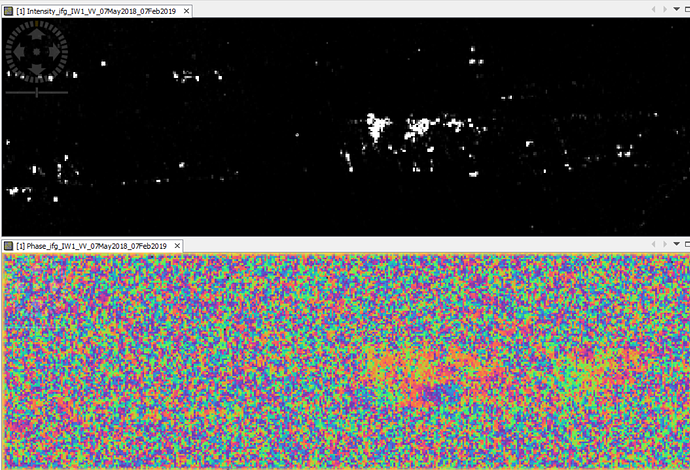
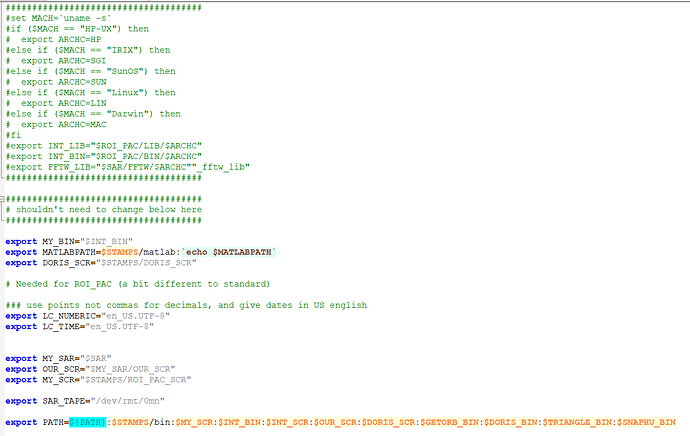
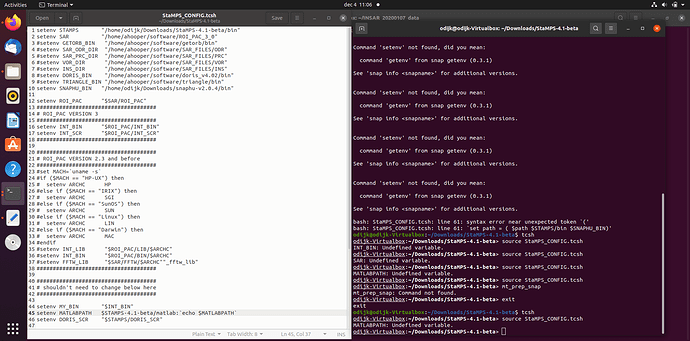
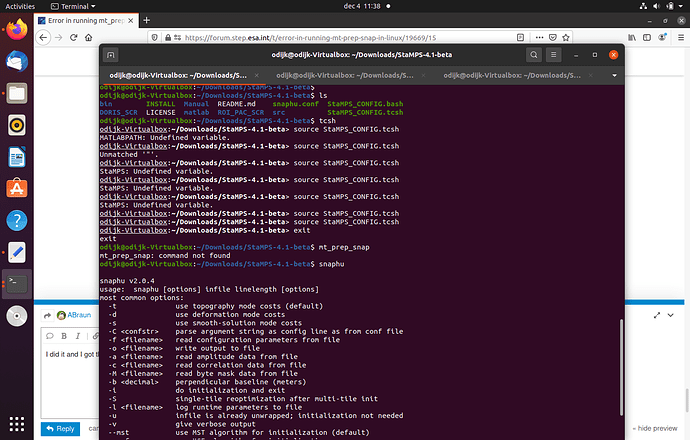
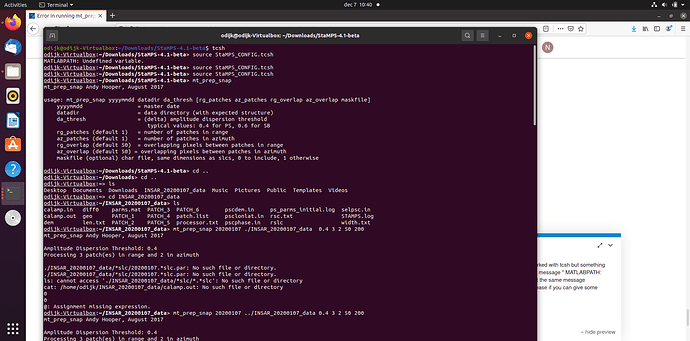
Hi, everyone. I am trying to use the script"mt_prep_snap" but I meet some problem. Please help me .
ubuntu@ubuntu-Precision-3630-Tower:~/Desktop/6TB/bodi/application$ mt_prep_snap 20190314 /home/ubuntu/Desktop/6TB/bodi/application/application/wokdir/PROC/INSAR_b.dim 0.4 3 2 50 200
csh: Input/output error
csh: Trying to start from “/home/ubuntu”
mt_prep_snap Andy Hooper, August 2017
Amplitude Dispersion Threshold: 0.4
Processing 3 patch(es) in range and 2 in azimuth
Segmentation violation detected at 一 12月 02 19:47:41 2019 +0800
Configuration:
Crash Decoding : Disabled - No sandbox or build area path
Crash Mode : continue (default)
Default Encoding : UTF-8
Deployed : false
Desktop Environment : ubuntu:GNOME
GNU C Library : 2.29 stable
Graphics Driver : Unknown software
MATLAB Architecture : glnxa64
MATLAB Entitlement ID : 5652177
MATLAB Root : /home/ubuntu/soft/matlab
MATLAB Version : 9.5.0.944444 (R2018b)
OpenGL : software
Operating System : Ubuntu 19.04
Process ID : 24954
Processor ID : x86 Family 6 Model 158 Stepping 10, GenuineIntel
Session Key : 64b20a93-480c-46ce-9323-745d1b1de2ce
Static TLS mitigation : Enabled: Full
Window System : No active display
Fault Count: 1
Abnormal termination
Register State (from fault):
RAX = 000000000000000b RBX = 00007f16ef998760
RCX = 787564642e62616c RDX = 00007f16ef998760
RSP = 00007f16ef998430 RBP = 00007f16ef998460
RSI = 0000000000000000 RDI = 00007f16ef998430
R8 = 0000000000000000 R9 = 0000000000000000
R10 = 00000000000000f0 R11 = 00007f171951d5d0
R12 = 00007f16ef998530 R13 = 00007f16ef998ec0
R14 = 00007f16ef998530 R15 = 00007f16e815d6d8
RIP = 00007f1711b36a22 EFL = 0000000000010206
CS = 0033 FS = 0000 GS = 0000
Stack Trace (from fault):
[ 0] 0x00007f1711b36a22 bin/glnxa64/libmwsettingscore.so+00973346 _ZNK8settings4core8Settings21getSettingsByFullNameERKNSt7__cxx1112basic_stringIcSt11char_traitsIcESaIcEEE+00000018
[ 1] 0x00007f16f5bb31e5 bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00725477
[ 2] 0x00007f16f5b830fb bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00528635
[ 3] 0x00007f16f5b6c500 bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00435456
[ 4] 0x00007f16f5b78eca bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00487114
[ 5] 0x00007f16f5b72eaa bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00462506
[ 6] 0x00007f17167cce75 bin/glnxa64/libmwms.so+02625141
[ 7] 0x00007f16f5b72b06 bin/glnxa64/ddux/ddux_impl/mwddux_impl.so+00461574
[ 8] 0x00007f171764ec0d /home/ubuntu/soft/matlab/bin/glnxa64/libmwboost_thread.so.1.65.1+00080909
[ 9] 0x00007f1716a77182 /lib/x86_64-linux-gnu/libpthread.so.0+00037250
[ 10] 0x00007f1717da5b1f /lib/x86_64-linux-gnu/libc.so.6+01170207 clone+00000063
[ 11] 0x0000000000000000 +00000000
** This crash report has been saved to disk as /home/ubuntu/matlab_crash_dump.24954-1 **
MATLAB is exiting because of fatal error
Killed
opening /home/ubuntu/Desktop/6TB/bodi/application/application/wokdir/PROC/INSAR_b.dim/rslc/20190206.rslc…
Segmentation fault (core dumped)
8013
2437
mt_extract_cands Andy Hooper, Jan 2007
Patch: PATCH_1
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij
Patch: PATCH_2
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij
Patch: PATCH_3
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij
Patch: PATCH_4
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij
Patch: PATCH_5
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij
Patch: PATCH_6
selpsc_patch /home/ubuntu/selpsc.in patch.in pscands.1.ij pscands.1.da mean_amp.flt f 1
file name for zero amplitude PS: pscands.1.ij0
dispersion threshold = 0.4
width = 24039
number of amplitude files = 0
Segmentation fault (core dumped)
psclonlat /home/ubuntu/psclonlat.in pscands.1.ij pscands.1.ll
opening pscands.1.ij…
Error opening file pscands.1.ij
pscdem /home/ubuntu/pscdem.in pscands.1.ij pscands.1.hgt
opening pscands.1.ij…
pscdem: Error opening file pscands.1.ij
pscphase /home/ubuntu/pscphase.in pscands.1.ij pscands.1.ph
opening pscands.1.ij…
Error opening file pscands.1.ij