I run a RGB composition and now I want to run the Biophysical Processors for LAI but this message appairs
Error: Operator Exception
Message Missing band at 560.0 nm
Anyone can help me?
I run a RGB composition and now I want to run the Biophysical Processors for LAI but this message appairs
Error: Operator Exception
Message Missing band at 560.0 nm
Anyone can help me?
what data are you using?
Hi, I’m using .GEOTIFF
SNAP needs the data in its original form. In case you process it somehow, make sure you select BEAM DIMAP as output format because it conserves all metadata required for such tasks.
A Geotiff file doesn’t contain any metadata, therfore the Biophysical Processor fails.
Thanks, but now I tried to run with the original data I got the same error…
I used the process bandmerge (RGB 843) and run the result of this process to run the biophysical process, it is right?
And, I now I can open the .zip file with all bands of sentinel but it is not working for me also. I have to open each band individualy and run the bandmerge…
actually, there is no merging needed for the biophysical parameter tools. The only thing important is that you have all metadata (calibration is also advisable).
What error do you get if you directly apply the tool on the original data?
I get the same error…
Error: Operator Exception
Message Missing band at 560.0 nm
If I send you the log do you think you can see anything else?
what sensor are you using and how did you load the data into SNAP?
I’m using SENTINEL2 data loaded from USGS
To load the data into SNAP, as I said before, I dont know why but I cant load the zipfile with all band of santinel, the SNAP said that this file is not approprieted product, Then I load by - File/ import/ GenericFormat/ JPG-2000. and import eatch dand as a new project. After to merge them to run the biophysical I used the Graphbuilder with bandmerge.
this is not the correct way to import Sentinel-2 data.
Make sure you have the latest version of SNAP (Menu > Help > Check For Updates) and try it like this:
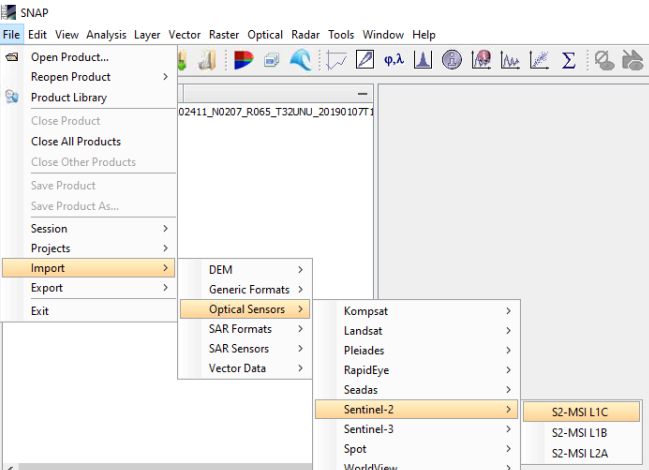
The data is then loaded as a full stack with all required metadata:
But as Sentinel-2 is a multi-size product, you have to resample it first before you can run the biophysical parameter estimation.
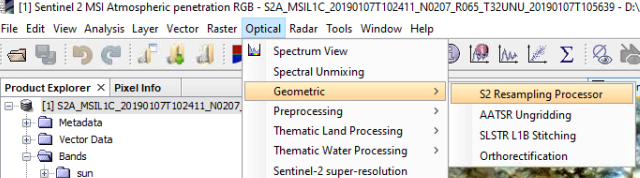
Ok thanks, I’m updating
By this way to load the data, witch tipe of data I load? the zipfile with all bands? or the metadatL1C?
sorry for not mentioning it in the first place.
You extract it and select the file that contains MTD
And I do the Resampling with the MTD also? Or with the image merged?
another doupt… To run the biophysical I have to merge the bands? And what bands? 843 is correct?
I dont know but I cauld not open the MSD, it get an erro of not appropriet product…
I’m gonna reinstal the SNAP…
yes, this is the input to any tools. If you open the MTD the bands are automatically used as a stacked product. No merging is needed.
I don´t know why… but this process is not working… do you have the link to my download the correct version of SNAP?
http://step.esa.int/main/download/
Did you see that installing version 6 is not enough? You also have to collect the internal updates (Menu > Help > Check for Updates)
Thank you so much, it is running now.
But I now I have to do some corrections on the SENTINEL image before run the biophysical, can I perform it in the SNAP?
how did you solve your problem?
For any automated retrieval of information, radiometric calibration is advisable. You can do it with the sen2cor plugin. It can be downloaded here: Sen2Cor – STEP
The easiest way is launching it from the command line as described here: Sen2Cor-02.05.05-win64 - AttributeError: 'L2A_Tables' Object has no attribute '_L2A_Tile_PVI_File' - #4 by Fabrizio_Ramoino