Thank you for replying @ABraun ,
i tried your said too. But i got same error again. Maybe it connected with Matlab version i use . i m using 19 version which is the latest released
Here might be the issue!!
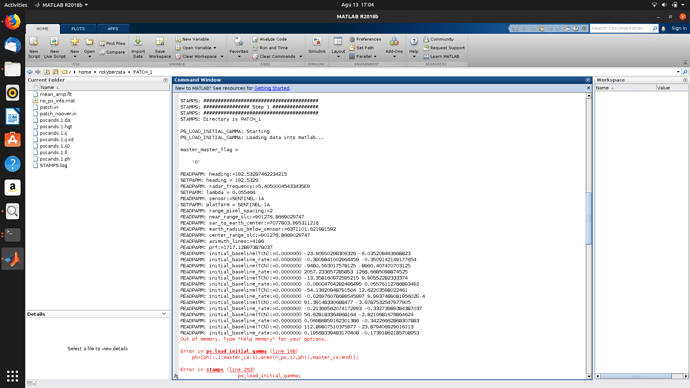
Lambda is NaN? With no lambda defined, cannot derive displacement from phase
as well as heading and the error message said cannot check if Envisat.
Could you please set up those parameters?
Which StaMPS version are you using?
This should be set up from early steps… step 1 . How did you manage to go through it without getting errors?
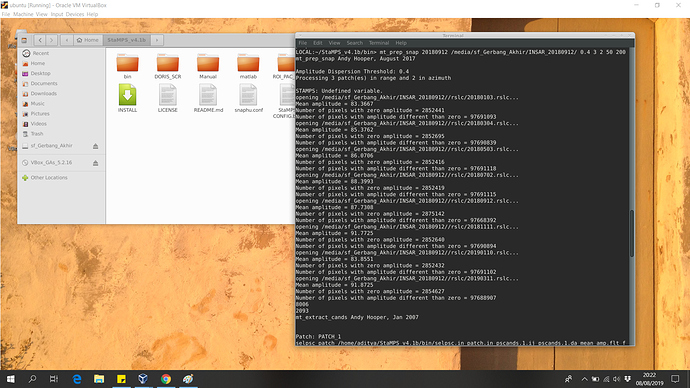
Hello everyone, i’m using oracle virtualbox to run my Linux Ubuntu OS.
I have installed StaMPS v.4.1b.
I’m currently having a problem with mt_prep_snap
Please take a look at my terminal below:
Best regards.
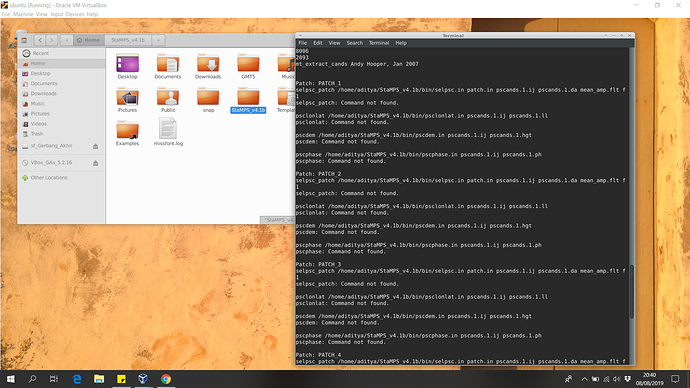
Please include the path where those binaries are which should be included when you load the StaMPS_CONFIG.bash
It seems that the running inside that folder does not recognized the other functions so be sure you include that path on the enviromental PATH variable or similar
Dear @mdelgado
Actually NaN doesn’t mean that ‘null’ , ‘0’ or ‘unnadjucted’. It refers ‘default’ and i think there is no obligation to change it exactly.
I have solved the problem that has forced me for a few days. I can say some hints to persons who may face with this kind of problem after completing all steps at matlab with StaMPS during plot processing. The most important thing is sequence of process.
1-Close the matlab and terminal when you see this error How to prepare Sentinel-1 images stack for PSI/SBAS in SNAP
2-Reprocess mt_prep script in the terminal
3-Run the matlab after mt_prep script in the terminal complete succesfully
4-If needed change entering setparm(scla_deramp…) and setparm(insar_processor)
5-Start the process in matlab typing stamps(,) step you want up to complete all 8 steps
6-After completing all 8 steps succesfully if needed setparm(centre_lonlat……) and setparm(ref_radius…)
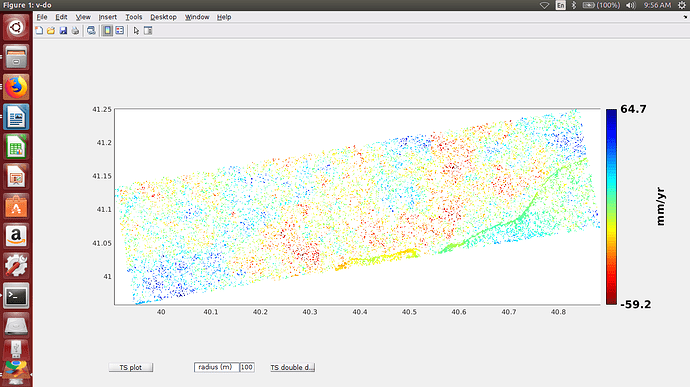
7-Now can be displayed what you want to plot (see example below).
Best regards
Ketti
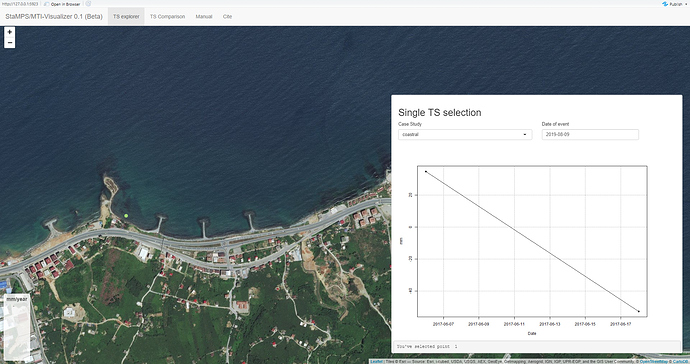
i met something weird situation using with StaMPS_Visualizer 0.1 (Beta). After i exported csv file from the matlab, only 1 PS point is displayed. Could you say if i have to change smth. in standart code below or matlab process before export csv file .( i did not face any problem both exporting and displaying as kml format) Thank you.
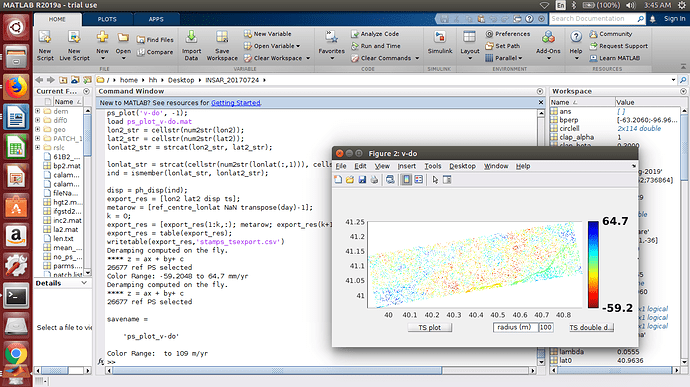
ps_plot(‘v-do’, ‘ts’);
load parms.mat;
ps_plot(‘v-do’, -1);
load ps_plot_v-do.mat
lon2_str = cellstr(num2str(lon2));
lat2_str = cellstr(num2str(lat2));
lonlat2_str = strcat(lon2_str, lat2_str);
lonlat_str = strcat(cellstr(num2str(lonlat(:,1))), cellstr(num2str(lonlat(:,2))));
ind = ismember(lonlat_str, lonlat2_str);
disp = ph_disp(ind);
export_res = [lon2 lat2 disp ts];
metarow = [ref_centre_lonlat NaN transpose(day)-1];
k = 0;
export_res = [export_res(1:k,:); metarow; export_res(k+1:end,:)];
export_res = table(export_res);
writetable(export_res,‘stamps_tsexport.csv’)
Dear @mdelgado
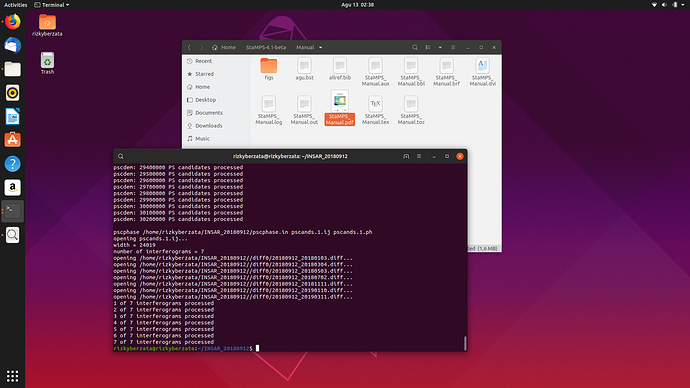
Thank you for helping me in the process of making mt_prep_snap.
Yesterday, I installed Linux Ubuntu.
below is my mt_prep_snap result:
then I open matlab and enter the command stamps (1.1)
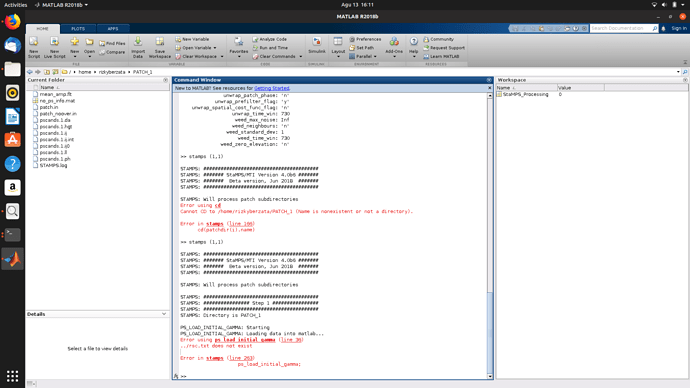
then I get an error “cannot cd to / home / rizkyberzata / PATCH_1”
so I copy my PATCH_1 folder which was previously in the “INSAR_20180912” folder to “/ home / rizkyberzata”
I run the command 1.1 again.
I get an error “…/rsc.txt does not exist” in ps initial load gamma
so I copy everything in the “INSAR_20180912” folder to "home / rizkyberzata except for the “dem, diff0, geo, rslc” folder.
then I get an error “out of memory”
Best regards.
I think both can be handled by the correct use of mt_prep_snap:
- I can recommend to create a working directory first
(at a suitable location, e.g. home\rizkyberzata \20180912_work) and start the script in there. Accordingly, all newly generated files are created in this folder. This not only helps you to keep it clean, but also allows you to quickly delete all required files in case you want to start again without missing a file. - If you have memory issues, you can increase the number of patches.
mt_prep_snap DATE location\of\preprocesed\data 0.4 2 2 50 200
This creates four PATCH folders which automatically split your area into smaller pieces. Matlab is later able to handle this structure and requires less memory for the processing of single steps.
I see the issue that you got a different path when you have run the mt_prep_snap inside the INSAR_20180912 and later inside matlab you do not get the same path tree…
mt_prep_snap saves the full path to some files, so please replicate the same folder tree and run matlab commands again. it should work
Using matlab you can always limit the points based on coordinates, velocities, height… etc at your wish.
Okay, I have finished Stamps (8,8).
Now, before I visualize the results, I want to limit the area that will be displayed, how to do it?
What command should I use? and would you give an example of the number coordinate format entered in the command?
Thanks in advance 
Hi @berzata,
If you read StaMPS manual or the ps_plot usage you will find what you are looking for.
No need to start everything from scratch. I think these are good news for you
Some of the plotting options 
BACKGROUND = -1 outputs the data to a .mat file instead of plotting
0, black background, lon/lat axes
1, white background, lon/lat axes (default)
2, shaded relief topo, lon/lat axes
3, 3D topo, lon/lat axes
4, mean amplitude image
5, mean amplitude image, brightness showing through PS
6, white background, xy axis (rotated lon/lat)
PHASE_LIMS = 1x2 vector with colormap limits (or 0 for default)
defaults to the range of the plotted phase
REF_IFG = number of interferogram to reference to - defaults to 0 (master)
-1 for incremental referencing
IFG_LIST = list of interferograms to plot - defaults to [] (all)
N_X = maximum number of images to plot per row
defaults to 0 (find optimum based on image size)
CBAR_FLAG = colorbar flag - defaults to 0 (plot on master, if plotted)
1 = don't plot a colorbar
2 = plot a colorbar underneath
TEXTSIZE = size of date text in points - defaults to 0 (best)
+ve size plots a top (default), -ve size plots at bottom
TEXTCOLOR = 1x3 color vector - default white or black depending on BACKGROUND
LON_RG = longitude range - defaults to [] (whole image)
LAT_RG = latitude range - defaults to [] (whole image)
'ts' = produce time series plot on user click over velocity plots.
the position of this switch is not important.
Note needs to be SM (v-option) of SB inverted to SM
time-series (V-option)
'ifg i' = only for 'a_p' show the topopgraphy correlated aps correction
for the ith interferogram for all spatial bands. Note that
the definition of ifg_list changes the spatial bands for this option.i also met the problem in step 1, and i delete all the patch file and regenerate patches and do step 1, and it works
Hello everyone
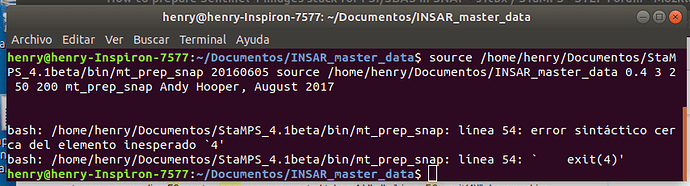
I have a problem with mt_prep_snap, I am working with ubuntu linux and stamp 4.1b I have the files exported from SNAP:
bash: /home/henry/Documentos/StaMPS_4.1beta/bin/mt_prep_snap: línea 54: error sintáctico cerca del elemento inesperado `4’
bash: /home/henry/Documentos/StaMPS_4.1beta/bin/mt_prep_snap: línea 54: ` exit(4)’
please, thanks for your input
I strongly believe that the problem is in the way you run mt_prep_snap
Please check the command used there… not sure why you use a source after the master date and the insar folder…
The problem is indeed in how you run the commands.
- first Source the stamps-config file (this will create the correct environment and packages for stamps)
- then run the mt_prep_snap command as shown in the StaMPS manual.
- for more instructions you can check the readme in my thesis github:
Greetings,
Gijs
Gracias: Intentare nuevamente
thank you, I will interact again