Hi everyone, I have the following problem to solve.
I save as .tif a sentinel-2 observation, import it in Python (using gdal), extract the top canopy reflectance bands (B1,…,B12), and compress them by using a certain compression method, which outputs an approximation of these bands (B1_approx,…,B12_approx).
Here the problem.
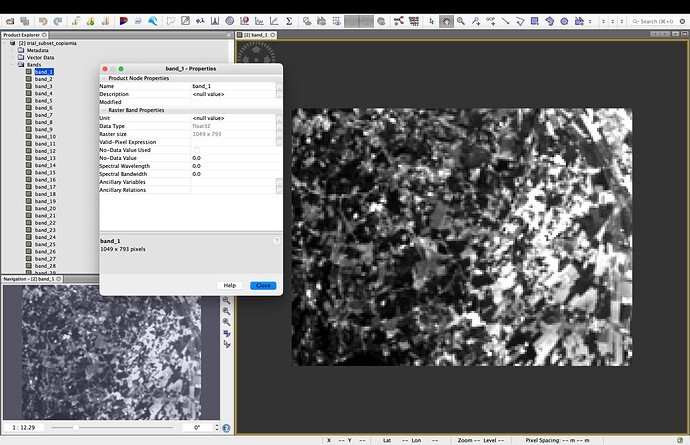
I would like to compute the LAI starting from the reconstructed observation, but in order to perform this task, I should have to build another optical observation replacing the exact bands with the reconstructed ones, and save it as .tif. I tried but maybe I’m not performing correctly the task, because, when I compute the LAI in SNAP starting from the reconstructed observation, I get an error saying me that the software is not able to recognize the bands. Indeed, in the .tif where I have replaced the original bands with the reconstructed ones, the name of the bands are different and also the structure is different.
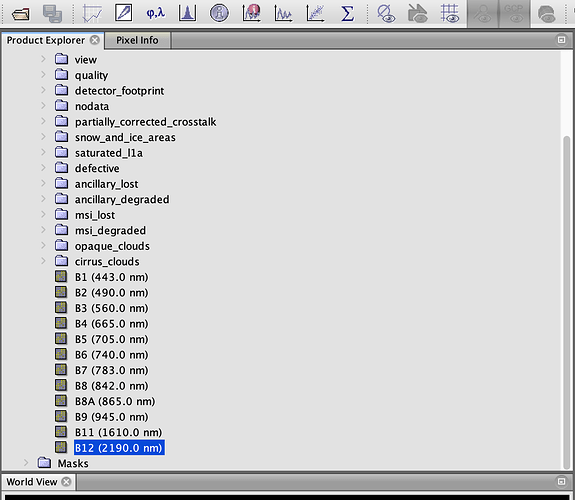
I don’t see the traditional structure (see attachment) but the list of all the bands (also ancillary data), without subfolders as in the original case.
Can anyone help me? Summing up, the actual problem is the following: I am not able to replace some bands in an optical observation so that SNAP is able to read correctly the modified .tif.
I am getting crazy… ![]() Thanks in advance.
Thanks in advance.