you can simply delete all downloaded folders.
I found the problem.
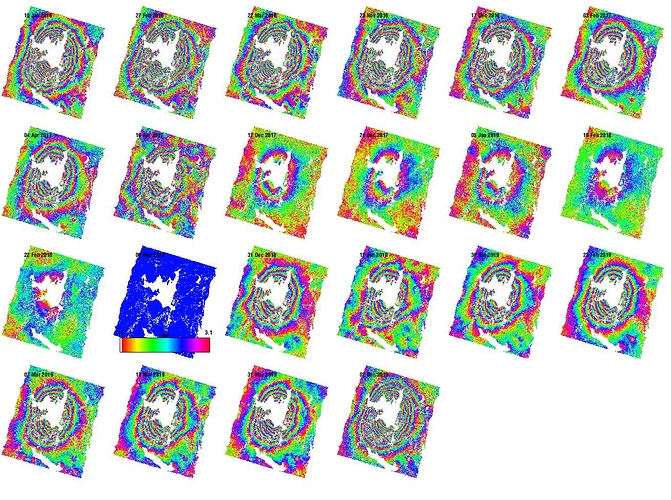
It was my fault for creating one of the xml routines (I modified them to have more intermediate steps). By doing so I changed the order of the master-slave and this error was dragged into the StaMPs. here is the w phase with some dmo errors remove.
Thank you very much for your help and attention.
Perfect!!!
Really nice results! I look forward to read your paper!
Could you please let me know what happened in your study area? Which event has created these fringes there?
I am running all this process in a virtual os were I installed matlab. How can I save the workspace to a stamp step if I want to close and view the results later?
any step you have processed is already stored in the files in your working directory. Simply open matlab again in the same directory and you can continue where you have finished last time.
Or do you mean how to export the graphs? https://de.mathworks.com/help/matlab/creating_plots/saving-your-work.html
thank you so much, all I had to do was replacing the code in stamps matlab folder and then rerun stamps (1,1) , right?
I’m facing it again:
provide more information… where are you running the script? how?
can you check if you got empty files in the PATCH_XXX folders?
Otherwise it is difficult to figure out the issue
I opened matlab in the folder I exported the data from snap and used “mt_prep_snap” in. then I had run stamps(1,1) and faced the error
oops! yes these there files are 0 bytes in patch1 folder , and also I don’t have path2 in folders :-?
how should I fix it now?
You should check the interferograms and check whether some of the slaves produced an empty / partially empty interferogram.
Open the products in SNAP and check them. Also the coregistered stack.
My guess is that something went wrong before the mt_prep_snap.
Check the calamp.out to see if any slave had 0 amplitude average.
Thank you for the suggestion about calamp.out.
If I have got a slave with 0 amplitude, will it be better to re-do all the snap processing?
not necessarly:
you can:
- re-do that slave processing (split, coreg_ifg, export, stamps_export) and the full mt_prep_snap
- remove it slave from the rslc and pair master-slave ifg from the diff0 folder (within the INSAR_XXXX folder) and re-do the full mt_prep_snap
Let us know
Hi,
I re-did all the snap processing to be sure but a slave remains with 0 amplitude. So I deleted it and I run again mt_prep_snap and now everything is ok.
Thanks again
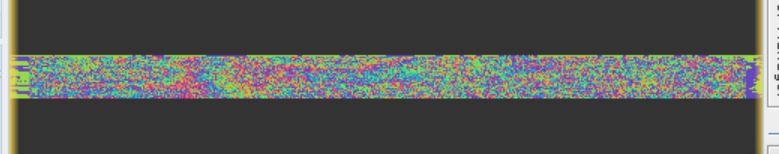
this is the phase of topophse removal step:
is there anything wrong with these?
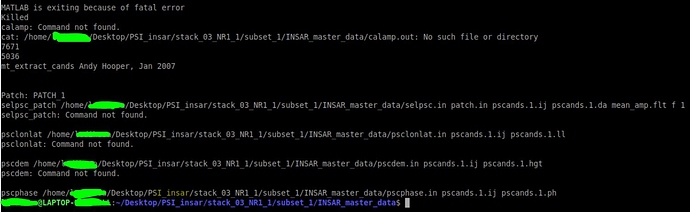
I did the mt_prep_snap for just one burst and after that i got 6 patch folders, so the run went better than the previous time but I also faced these, all files where ok but all the ij0 files where 0bytes this time:
!
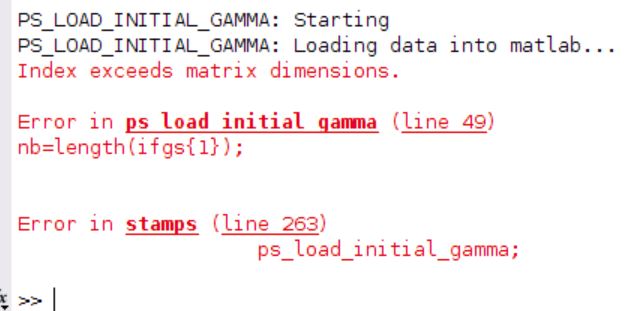
I face this when i open matlab:
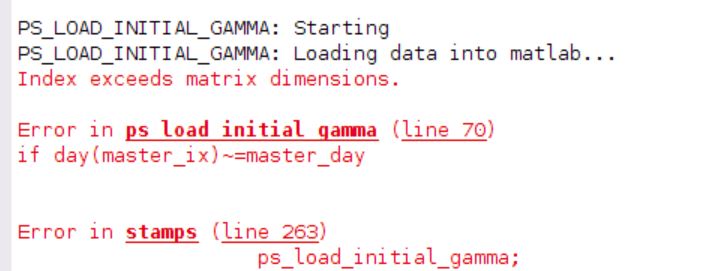
Hello, i had the same problem , with the matlab error stamps (2,2) . so i tried to change the function ps_load_initial_gamma.m as you said. now i have a bigger problem, as the mt_prep_snap command doesn’t end well. I tried to change the function with the backup file, but it is the same result as below:
had you added the mt_prep_snap command into the PATH environment? Normally you get it done by sourcing the StaMPS_CONFIG.bash
let me know if this helped.
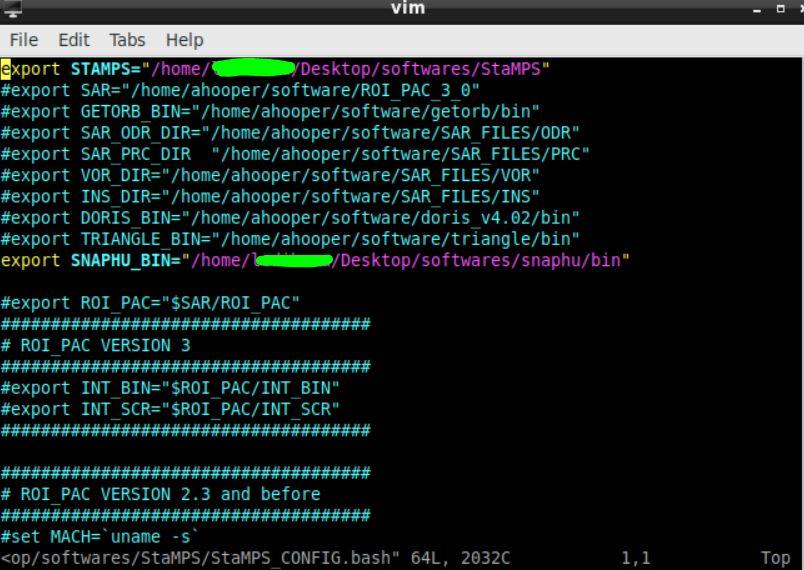
yes, I did, the first time I run it without changing the ps_load_initial_gamma.m , it worked. now it seems to have a problem. maybe the snaphu path folder in the StaMPS_configure.bash is wrong. How do i find out where to extract the path? I have extract it in windows and then installed it from ubunutu commands. This is how my file .bash look like
Let me just say that ps_load_initial_gamma.m file has no relation to the mt_prep_snap.
The first runs in the stamps step 2 while the mt_prep_snap runs before stamps step1.
If you have changed environment maybe this is the problem.
had you successfully compiled the stamps/src folder in your new environment?
if so, which gcc version you have used? there are threads about it as well.
best
i’m sorry, gcc version is the version of the StaMPS ? if so, i’m using the stamps 4.1b
no. gcc is the c compiler you should had used to compile the src folder provided within the stamps folder.