Hi i’m working with snappy and my scripts works so well until when i arrive at 2010 file. After that moment in my terminal apears the error:
Traceback (most recent call last):
File “./complete_output_file.py”, line 67, in
ProductIO.writeProduct(result,outDir,outForm)
RuntimeError: java.lang.NullPointerException
My script is:
#!/usr/bin/python
import sys
import glob
#import netCDF4
from netCDF4 import Dataset as ncDataSet
import snappy
from snappy import ProductIO
from snappy import GPF
import numpy as np
import os
# region
if len(sys.argv) >= 1:
region = sys.argv[1]
else:
sys.exit("Incorrect number of arguments for " + sys.argv[0])
# TOA path files
path=region + '/data/*'
list=sorted(glob.glob(path), key=os.path.getmtime)
#list=glob.glob(path)
for e in range(len(list)):
# open netcdf file to know TOA file
ncFile=ncDataSet(list[e],'a',format='NETCDF4')
TOA=ncFile.__dict__['l1_file']
# Just execute if file is not processed
if 'sst_nadir' in ncFile.__dict__ :
nom=TOA[17:76]
# SNAP - Optical - Thematic water Processing - (A)ATSR SST Processor
source=ProductIO.readProduct(TOA)
op_spi=GPF.getDefaultInstance().getOperatorSpiRegistry().getOperatorSpi('Aatsr.SST')
param_Desc=op_spi.getOperatorDescriptor().getParameterDescriptors()
#for param in param_Desc:
HashMap=snappy.jpy.get_type('java.util.HashMap')
parameters=HashMap()
parameters.put("dual","false")
parameters.put("nadir","true")
parameters.put("nadirCoefficientsFile","AVERAGE_POLAR_SINGLE_VIEW")
parameters.put("nadirMaskExpression","!cloud_flags_nadir.LAND")
parameters.put("invalidSstValue","-999.0")
result=snappy.GPF.createProduct('Aatsr.SST',parameters,source)
#outDir='/mnt/lustre/scratch/'+nom+'.nc'
outDir='/home/masdeu/'+nom+'.nc'
outForm='NETCDF4-CF'
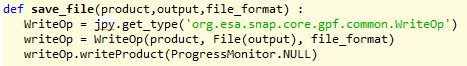
ProductIO.writeProduct(result,outDir,outForm)
start=ncFile.__dict__[u'start_line']
end=ncFile.__dict__[u'end_line']
ncFile2=ncDataSet(outDir,'r',format='NETCDF4')
sstCor=ncFile2.variables['nadir_sst']
lati=ncFile2.variables['latitude'][:]
loni=ncFile2.variables['longitude'][:]
A=sstCor[start:end+1,:]
B=lati[start:end+1,:]
C=loni[start:end+1,:]
# TOA processed result has a mirror effect, for that reason we use a fliplr command before save the result in the final file
E=np.fliplr(A.data)
lat_TOA=np.fliplr(B)
lon_TOA=np.fliplr(C)
#sst_cell=ncFile.variables['sst'][:]
lat_cell=ncFile.variables['lat'][:]
lon_cell=ncFile.variables['lon'][:]
#rmse between ghrsst and toa
rmse_lat=np.sqrt(((lat_TOA-lat_cell)**2).mean())
rmse_lon=np.sqrt(((lon_TOA-lon_cell)**2).mean())
if rmse_lat>=0.005 or rmse_lon>=0.005:
print('ATENTION! LAT-LON DISTANCE>=500m')
ncFile.rmse_lat = rmse_lat
ncFile.rmse_lon = rmse_lon
else:
ncFile.rmse_lat = rmse_lat
ncFile.rmse_lon = rmse_lon
# Write sst values from Corrected TOA to original netCDF
ncFile.variables['sst'][:]=E
del ncFile.sst_nadir
write=list[e] + ' Complete'
print(write)
ncFile.close()
ncFile2.close()
os.remove(outDir)
print("File Removed!")
else:
write=list[e] + ' SST added'
print(write)
you have some idea?
thank you!