I want to remove clouds from Sentinel-2 imagery (Level 2A) using ESA SNAP. Please guide me.
I use SNAP 9.0.0.
I need the imagery to be cloud free since I am carrying out a land use land cover classification.
If you use L2A data, you already have a cloud mask available which you can use to exclude those pixels from further processing.
See here:
sen2cor is the processor used to generate L2A products.
Thank you for your support.
My file is “S2A_MSIL2A_20220929T050611_N0400_R076_T44NMM_20220929T073656”. Please find my file attached herewith.
I want to remove clouds and shadows in here. Can you please guide me to solve this matter.
Hi,
I want to remove clouds on Sentinel-2 Level 2A data. I want to use the scl_cloud_*_proba flags in the quality_scene_classification flag band or the value of quality_cloud_confidence to remove them.
I don’t have any idea to proceed this.
Please guide me.
Hi,
I’ve moved latest thread you’ve created to this one.
You can do experiments with the flags either with the Band Maths or with the Mask Manager.
For both exists tutorials on youTube.
SNAP - Band Math - YouTube
SNAP - Mask Manager - YouTube
Unfortunately, both have a terrible voice over. But they show how to work with it.
With both you can find the expression you would like to use for masking the clouds.
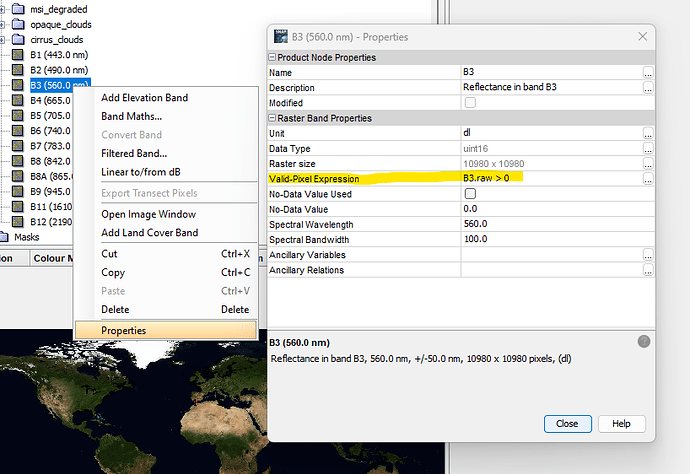
Once found you can use it as valid-pixel expression in the bands used for the RGB.
You can set this expression in the properties dialog.
Thank you for your support.
After that I want to fill the removed clouds with another (same tile number T44NNN).
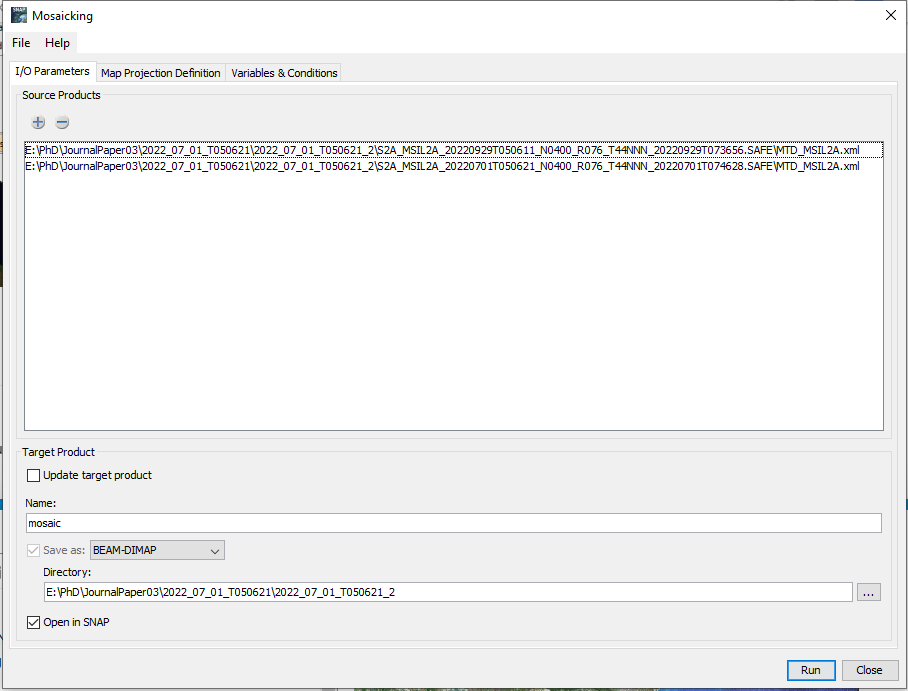
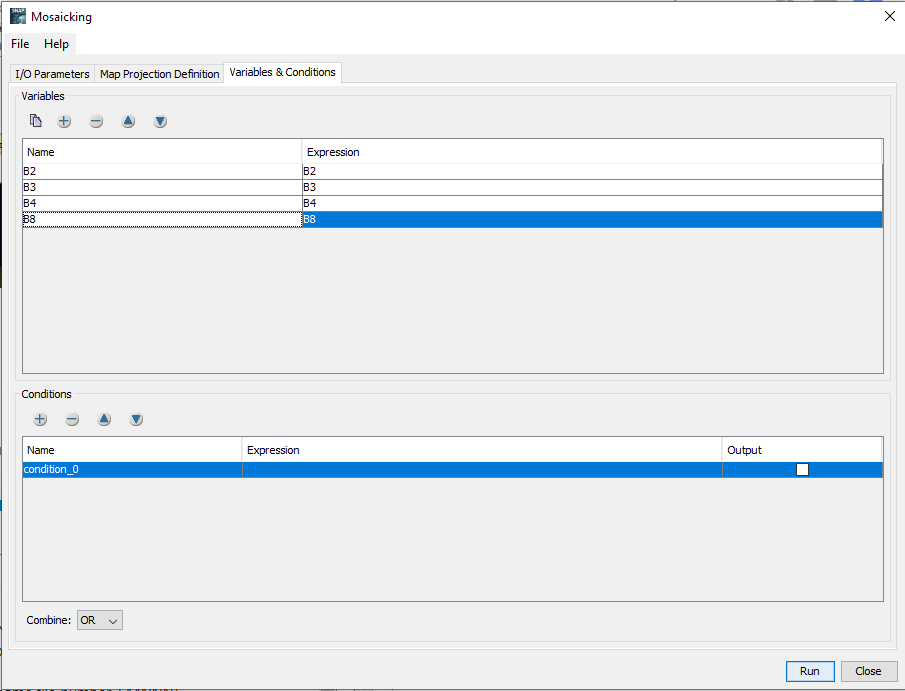
I followed the path Raster<<Geometric<<Mosaicking
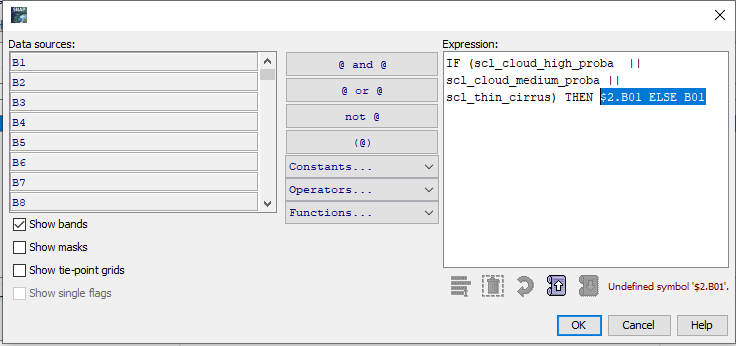
I noticed your answer in Cloud mask - SNAP sentinel 2. I tried to use as expression the following :
IF (scl_cloud_high_proba || scl_cloud_medium_proba || scl_thin_cirrus) THEN $2.B01 ELSE B01
But I couldn’t find $2.B01. I have only B1.
Please guide me to solve this matter. If I need to follow another method to solve my matter please help me.
The expression you have used is not working for the condition. In the end it must evaluate to true or false.
In the example Cloud mask - SNAP sentinel 2 - #52 by marpet I’m simply generating a new band by using the Band Maths. IN this case Mosaic is not necessary.
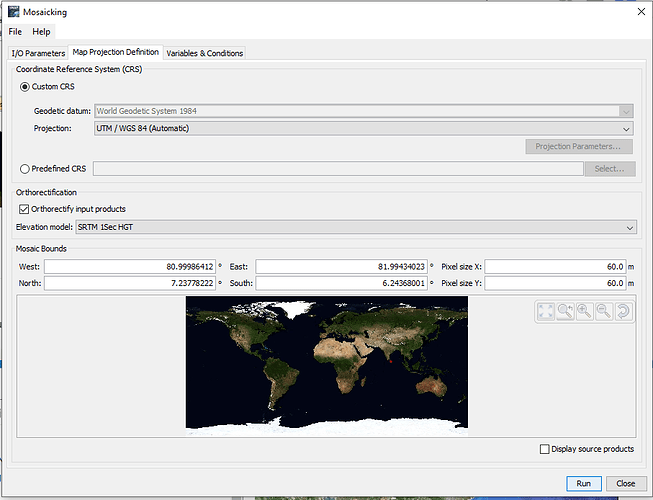
For the Mosaic condition you can use an expression like
not (scl_cloud_high_proba || scl_cloud_medium_proba || scl_thin_cirrus)
And you should not enable the orthorectification. Sentinel-2 L1C and higher is already orthrectified.
Thank you for your fast reply.
I’m currently following this procedure. But I have to face few issues.
- I resampleted my Sentinel-2 image
- Then resampled image Bands modified (B2.raw > 0 && !scl_cloud_medium_proba && !scl_cloud_high_proba)
- Mosaic the resampled modified image with other resampled image (Same tile number)
After this process I closed and save files. Then when I tried to resample the original Sentinel-2 image I received this error message.
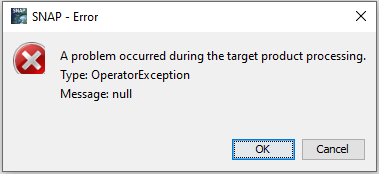
I tried to solve. It solved after I detele all files and extract files again.
Please give me your valuable suggetion regarding this matter.
Thak you.
Could you provide the log file please?
How to find it is described here: How to find the log files?
Thank you very much for your support. Please find the log files attached herewith.
messages.log (159.8 KB)
messages.log.1 (86.9 KB)
messages.log.2 (79.6 KB)
updater.log.0 (749.8 KB)
Thank you.
I’ve seen multiple issues in the logs.
One is that you are running out of disk space on drive E.
Caused by: org.esa.snap.core.gpf.OperatorException: Not able to write product file: 'E:\PhD\JournalPaper03\2022_09\S2A_MSIL2A_20220929T050611_N0400_R076_T44NMM_20220929T073656_resampled.dim'
at org.esa.snap.core.gpf.common.WriteOp.computeTile(WriteOp.java:375)
at org.esa.snap.core.gpf.internal.OperatorImage.computeRect(OperatorImage.java:82)
at javax.media.jai.SourcelessOpImage.computeTile(Unknown Source)
... 4 more
Caused by: java.io.IOException: There is not enough space on the disk
The other is:
Caused by: java.nio.BufferUnderflowException
at java.nio.Buffer.nextGetIndex(Buffer.java:506)
at java.nio.DirectByteBuffer.getInt(DirectByteBuffer.java:681)
at org.esa.snap.lib.openjpeg.dataio.RasterUtils.readAsByteArray(RasterUtils.java:70)
at org.esa.snap.lib.openjpeg.dataio.OpenJP2Decoder.decompress(OpenJP2Decoder.java:317)
at org.esa.snap.lib.openjpeg.dataio.OpenJP2Decoder.read(OpenJP2Decoder.java:142)
at org.esa.snap.jp2.reader.internal.JP2TileOpImage.computeRectDirect(JP2TileOpImage.java:133)
This happens when reading the S2A_MSIL2A_20220929T050611_N0400_R076_T44NNN_20220929T073656.SAFE product.
Maybe the product is broken. Have you tried other products?
Thank you very much for your support.
I have “S2A_MSIL2A_20210918T042711_N0500_R133_T46RDU_20230113T034810.SAFE” and “S2A_MSIL2A_20210928T042711_N0500_R133_T46RDU_20230112T220710.SAFE” sentinel 2 L2A images of same area. I used the expression in band math {if not (scl_cloud_shadow || scl_cloud_medium_proba || scl_cloud_high_proba || scl_thin_cirrus) then B4 else NaN} to extractred cloudy pixels from one image. Now how I can composite with other image to fill gaps. How to perform this task??
This can be achieved with the Mosaic operator respectively the multi-size Mosaic operator.
Set the expression as condition or include it in the variable definition.
You can also consider using the S2GM service.