I have come back to use snappy after some time, and I am seeing a slower processing of the snappy module when writing a product in a numpy array (10390x10390 pixels) , and also when trying to display a band with matplotlib , it do needs quite a longer time than before. Is this something common or it could only be something related to my system ( but I have not found any problem yet with it)
As far as I know there was no major change on the S2 Reader. Also not on snappy. I think you are using the latest SNAP5.x version, right?
yes, I keep SNAP updated to the latest release. Actually the critical operation is to display the image 10980x10980 pixels with matplotlib, it really needs a lot of time. Do you think it could be caused by the heavy size of the image?
Yes, maybe. I don’t use matplotlib So I can’t say if matplotlib is only efficient up to certain amount of data
Hello,
I have a question about the performance of snappy compared to the performance of SNAP toolbox.
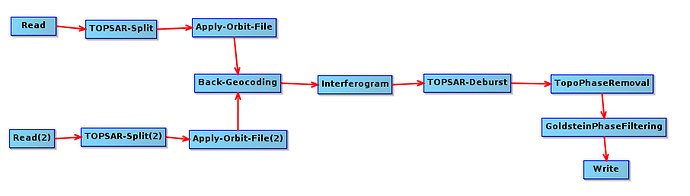
I am using SNAP toolbox for deriving a subsidence map. I decided to create a graph (the one that is shown below) to automate the procedure.
I also found convenient using the snappy in python for automating this procedure without having to start SNAP toolbox. The code i wrote is provided below. The code works perfectly fine but the only issue I encounter is the speed.
When I load the graph into SNAP, it takes no more than 1.5 hour for the whole process. On the other hand, when I run the code with snappy, it takes more than 4 hours. It does not make much sense to me because we are effectively accessing the JAVA modules using python as an interface. Hence, one expect to get results as fast as in SNAP toolbox
import os
from snappy import GPF
from snappy import ProductIO
from snappy import HashMap
from snappy import jpy
import subprocess
from time import *
# Hashmap is used to give us access to all JAVA oerators
HashMap = jpy.get_type('java.util.HashMap')
parameters = HashMap()
def read(filename):
return ProductIO.readProduct(filename)
def write(product, filename):
ProductIO.writeProduct(product, filename, "BEAM-DIMAP")
def topsar_split(product):
parameters.put('subswath', 'IW1')
parameters.put('selectedPolarisations', 'VV')
return GPF.createProduct("TOPSAR-Split", parameters, product)
def apply_orbit_file(product):
parameters.put("Orbit State Vectors", "Sentinel Precise (Auto Download)")
parameters.put("Polynomial Degree", 3)
return GPF.createProduct("Apply-Orbit-File", parameters, product)
def back_geocoding(product):
parameters.put("Digital Elevation Model", "SRTM 1Sec HGT (Auto Download)")
parameters.put("DEM Resampling Method", "BICUBIC_INTERPOLATION")
parameters.put("Resampling Type", "BISINC_5_POINT_INTERPOLATION")
parameters.put("Mask out areas with no elevation", True)
parameters.put("Output Deramp and Demod Phase", False)
return GPF.createProduct("Back-Geocoding", parameters, product)
def interferogram(product):
parameters.put("Subtract flat-earth phase", True)
parameters.put("Degree of \"Flat Earth\" polynomial", 5)
parameters.put("Number of \"Flat Earth\" estimation points", 501)
parameters.put("Orbit interpolation degree", 3)
parameters.put("Include coherence estimation", True)
parameters.put("Square Pixel", False)
parameters.put("Independent Window Sizes", False)
parameters.put("Coherence Azimuth Window Size", 10)
parameters.put("Coherence Range Window Size", 10)
return GPF.createProduct("Interferogram", parameters, product)
def topsar_deburst(product):
parameters.put("Polarisations", "VV")
return GPF.createProduct("TOPSAR-Deburst", parameters, product)
def topophase_removal(product):
parameters.put("Orbit Interpolation Degree", 3)
parameters.put("Digital Elevation Model", "SRTM 1Sec HGT (Auto Download)")
parameters.put("Tile Extension[%]", 100)
parameters.put("Output topographic phase band", True)
parameters.put("Output elevation band", False)
return GPF.createProduct("TopoPhaseRemoval", parameters, product)
def goldstein_phasefiltering(product):
parameters.put("Adaptive Filter Exponent in(0,1]:", 1.0)
parameters.put("FFT Size", 64)
parameters.put("Window Size", 3)
parameters.put("Use coherence mask", False)
parameters.put("Coherence Threshold in[0,1]:", 0.2)
return GPF.createProduct("GoldsteinPhaseFiltering", parameters, product)
def insar_pipeline(filename_1, filename_2):
print('Reading SAR data')
product_1 = read(filename_1)
product_2 = read(filename_2)
print('TOPSAR-Split')
product_TOPSAR_1 = topsar_split(product_1)
product_TOPSAR_2 = topsar_split(product_2)
print('Applying precise orbit files')
product_orbitFile_1 = apply_orbit_file(product_TOPSAR_1)
product_orbitFile_2 = apply_orbit_file(product_TOPSAR_2)
print('back geocoding')
backGeocoding = back_geocoding([product_orbitFile_1, product_orbitFile_2])
print('inerferogram generation')
interferogram_formation = interferogram(backGeocoding)
print('TOPSAR_deburst')
TOPSAR_deburst = topsar_deburst(interferogram_formation)
print('TopoPhase removal')
TOPO_phase_removal =topophase_removal(TOPSAR_deburst)
print('Goldstein filtering')
goldstein_phasefiltering(TOPO_phase_removal)
print('Writing final product')
write(TOPO_phase_removal, '/home/io/Desktop/test1.dim')
filename_1 = os.path.join('/home',
'io',
'Desktop',
'S1_SAR_files',
'S1A_IW_SLC__1SDV_20160609T224454_20160609T224521_011640_011CF1_B895.SAFE')
filename_2 = os.path.join('/home',
'io',
'Desktop',
'S1_SAR_files',
'S1A_IW_SLC__1SDV_20160727T224456_20160727T224523_012340_013377_67FF.SAFE')
insar_pipeline(filename_1, filename_2)
Is there anything particular in my code that might slow down the process? Do we expect snappy to be slower compared to SNAP?
It would be very useful to know why running snappy takes so much time for the process to complete.
I would appreciate any feedbacks
Using SNAP from snappy uses a different memory configuration.
Have you seen the section about memory configuration on this page: Configure Python to use the SNAP-Python snappy interface.
This might cause the slowness
HI marpet,
Thanks for your response. I opened the ‘snappy.ini’ file and set the ‘java_max_mem’ to 45GB.
My PC has 64GB of memory. Do you think if I increased the memory inside the snappy.ini file) to 64GB for instance, would that make the process faster?
No, I think the will not improve the performance anymore.
But maybe if you increase the size of the tile cache.
Change the property ‘snap.jai.tileCacheSize’ in snap.properties in the etc folder of the SNAP installation directory. A good value might be ~70% of the java_max_mem value
Thank you
I will try that
Johngan, this is the most beautiful piece of insar code I have seen in my entire life.
Thank you andretheronsa 
I have automated (pretty much) the whole procedure for producing a map subsidence using snappy.
I got stuck at the last step where we need to export the wrapped phase to snaphu format.
marpet, one of the developers kindly offered to help me with that by looking at my SAR data. Once this step is done, then i will upload the whole InSAR pipeline in this forum for everyone to use.
It will make our life easier 
Any news regarding this thread?
johngan, beautiful piece of code. Thanks for sharing!!!
Rafael
@johngan I’m running into issues with snappy as well. It’s slow and behaves differently from gpt+graph.
I wonder how you got everything running
Is your use case such that you cannot run gpt+graph?
Hello Markus!
Python is much nicer than bash to prepare the workflow.
Yes I could use gpt+graph but since you paid for the development of snappy go all the way and make it work 
You have plenty of users in this forum wanting to use Python and snappy.
Yes people clearly want to use Python and snappy and we need to improve this functionality. Still, there will always be a performance-hit with snappy, unless you run gpt via it.
First step would be to rename it, as Snappy is already the name of a google compression library and SnapPy, which is a package for studying the topology and geometry.
hi fabricebrito,
I understand what you mean. I just used snappy because i wanted to automate some tasks. I found it very useful but i had issues with performance (hopefully, you do not run snappy on a jupyter notebook as it gets even slower).
To be honest, i gave up on snappy and used GPD and bash script instead and everything works just fine.
I wish we did not have such performance issues with snappy as the majority of people prefer using python.
Hello All,
I have a question related to this thread, trying to do some SAR processing based on @johngan 's nice script but getting the following error message:
Traceback (most recent call last):
File "D:\my_project\graph\samples\sar_processing.py", line 49, in <module>
sar_pipeline(filename)
File "D:\my_project\graph\samples\sar_processing.py", line 43, in sar_pipeline
write(sar_pipeline, 'D:\my_project\graph\samples\test1.dim')
File "D:\my_project\graph\samples\sar_processing.py", line 19, in write
ProductIO.writeProduct(product, filename, 'BEAM-DIMAP')
RuntimeError: no matching Java method overloads found
What am I missing here?
The script I am running is:
import sys
sys.path.append('D:\\python\\Lib\\site-packages')
import os
from snappy import GPF
from snappy import ProductIO
from snappy import HashMap
from snappy import jpy
import subprocess
from time import *
#Hashmap is used to give us access to all JAVA oerators
HashMap = jpy.get_type('java.util.HashMap')
parameters = HashMap()
def read(filename):
return ProductIO.readProduct(filename)
def write(product, filename):
ProductIO.writeProduct(product, filename, 'BEAM-DIMAP')
def land_mask(product):
parameters.put('shorelineExtension', '20')
return GPF.createProduct("Land-Sea-Mask", parameters, product)
def calibration(product):
parameters.put("outputSigmaBand", True)
return GPF.createProduct("Calibration", parameters, product)
#Add Ellipsoid Correction here
def sar_pipeline(filename):
print('Reading SAR data')
product = read(filename)
print('Land Sea Masking')
product_Mask = land_mask(product)
print('Applying radiometric calibration')
product_Mask_Calib = calibration(product_Mask)
print('Writing final product')
write(sar_pipeline, 'D:\\my_project\\graph\\samples\\test1.dim')
filename = os.path.join('/my_project',
'graph',
'samples',
'subset.dim')
sar_pipeline(filename)
Any contribution will be appreciated.
E.
Found some mistakes and edited the code. Should be running now.