i have come to the STAMPS processing part, i got this error during stamps(2,2), do you know what it is?
You may like to share more information about which error you got on stamps step 2, otherwise we cannot blindly help you.
Can you be more specific?
Had you used the modified script I shared on another thread to be used in stamps step1?
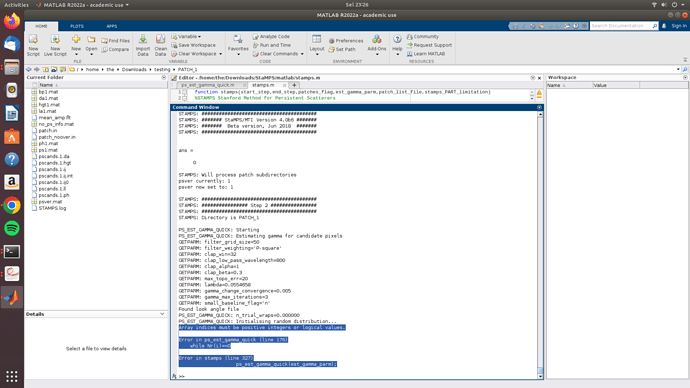
Array indices must be positive integers or logical values.
Error in ps_est_gamma_quick (line 176)
while Nr(i)==0
Error in stamps (line 327)
ps_est_gamma_quick(est_gamma_parm);
this is my error sir
looking at the n_trial_wrap=0.00000000 I believe that your IFG processing did not go well and at least 1 interferogram or secondary resampled image is not filled with right values.
i have problem with the stamps(2,2), the error is as below:
Error in clap_filt (line 47)
B=gausswin(7)*gausswin(7)';
Error in ps_est_gamma_quick (line 225)
ph_filt(:,:,i)=clap_filt(ph_grid(:,:,i),clap_alpha,clap_beta,n_win0.75,n_win0.25,low_pass);
Error in stamps (line 327)
ps_est_gamma_quick(est_gamma_parm);
Do you know what is the problem?
hello sir, I want to ask something, I got the result for my deformation, but the PS POINT did not lies on the coordinate for my monitored stations using GNSS as validation, is there a possible chance for me to improve my processing inside stamps??
you could apply less aggressive weeding in step 4 to keep more points in the analysis. But these are then potentially less correct.
Sir, i have problem, do you know during mt_prep_snap…why is there error that say “calamp.out not found”
hello sir, can you share me your processing parameter? if i can have your contact, will you give me?
hello sir, can you share me your processing parameter? if i can have your contact, will you give me?
please check the suggestions on weeding (step 4) here and especially here where the impact of the different variables is nicely explained.
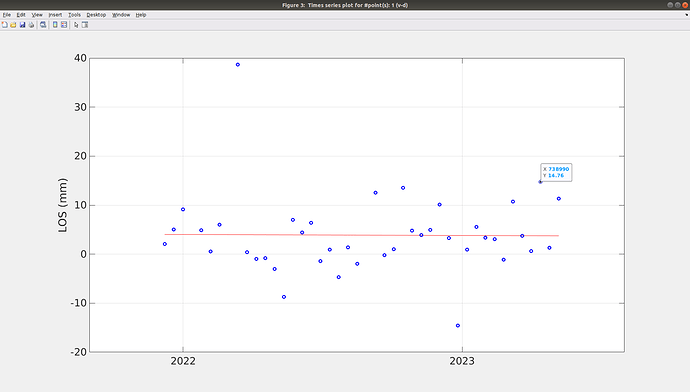
How do I know the actual z-value for the graph? because when i click on the point, it only shows the X Y value
Hi everyone,
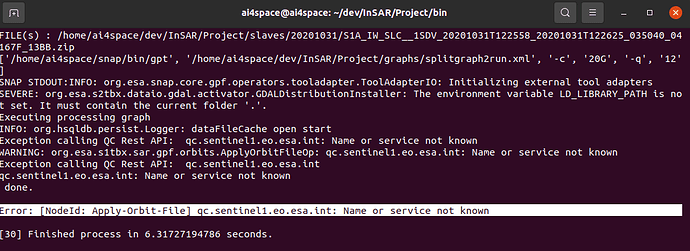
I have this error… the step splitting_slaves.py create the split folder and also the subfolders, but those are empty. The error is attached in the figure.
I am following the RUS webinar steps
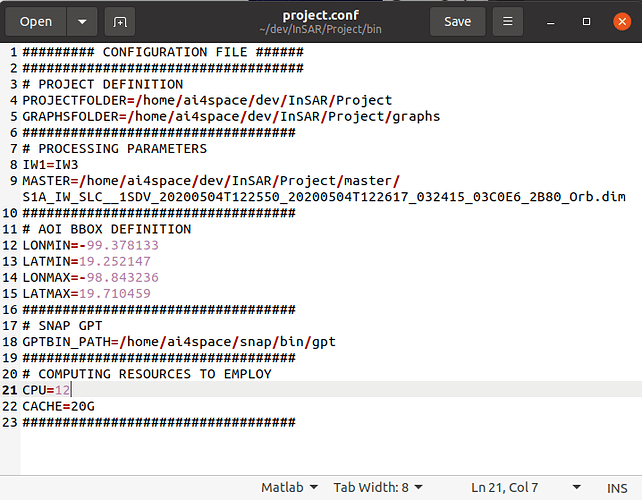
Please share the full project file.
I am using SNAP 6 and Python 2.7 as indicated by the tutorial, in order to have the correct compatibilities for SNAP2StaMPS conversion. My purpose is to extract persistent scatteres points
Have you split your master image (IW3)?
This is my project folder.
Yes, firstly I splitted the master by using SNAP application. Then I run slaves_prep.py, and now is the splitting_slaves.py turn.
If I run “python2 splitting_slaves project.conf”, in the current folder the split folder is created, in the same way also subfolders related to all the slaves but at the end they are empty. The error is the one that appear in the terminal.
I think that the problem is related to SNAP that is not able to connect to the server to download the orbit file, but if I upgrade SNAP i will have problems of incompatibility for SNAP2StaMPS conversion.
When I performed the split procedure for the master image i had the same problem, so i downloaded manually all the orbit files and the process ended successfully.
This version looks for the old address to get orbit files, you will have to update SNAP
You can pass already to snap2stamps v2 which uses SNAP v9