@ABraun if you don’t want to make .lat and .lon from stack_deburst. You can use .lat and .lon from Topo Phase Removal directly. But in coregistration, you must uncheck the mask area with no elevation.
I already try it and everything run smoothly
To make sure I’m not missing a key element or concept of the the solution by@reyhanrere that could cause problems later, I’m listing the steps I took that seemed to work below. If anyone sees anything that’s wrong let me know.
- Using Band Maths I created lat and lon bands in the stack_deb_subset product
- Using File>Export> I exported the product in Gamma format
- I now have lat.rslc and lon.rslc files (among other .rslc files for the other bands in the product)
- Renamed lat.rslc and lon.rslc files to .lat and .lon (ex 20170704.lat)
- Replaced the original 20170704.lat and 20170704.lon files in the geo folder with these new files.
- re-ran mt_prep_gamma (updated version)
- re-ran Stamps(6,6) (Didn’t think this would work but tried it anyway.)
Got a ‘matrix dimensions must agree’ error. - So I started from step one and ran Stamps(1,6) and the unwrapping completed successfully.
Thanks for looking it over.
By the way, any idea why the lat/lon files created with the SNAP6.0 ifg stage still need to be replaced in this manner?
I noticed that for some cases, the lat/lon files created in the StaMPS export work fine. I didn’t figure out what exactly causes the error at step 6.
Hi,
First of all, thank you for everyone, i got a lot of information about SAR processing by the STEP forum users. You are great! 
Thats my first try to do the whole SNAP-STAMPS PSInSAR process, sadly i stucked in the 6. step of stamps workflow.
I followed the recommendations of the official STEP topic (https://forum.step.esa.int/t/how-to-prepare-sentinel-1-images-stack-for-psi-sbas-in-snap-5), partly the MAINSAR google group topics/comments, and of course Thorstens StaMPS Workflow summary html.
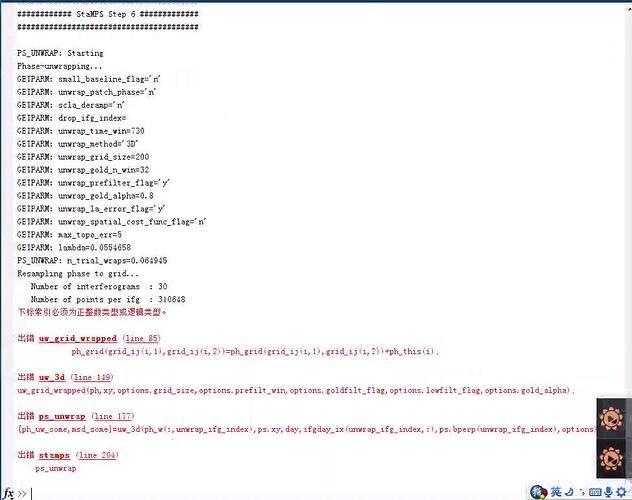
I got the following error in stamps step 6:
I tried the following things:
- Generate new lat and lon bands on back geocoded stack, then save to gamma format, and rewrite in the geo folder (from: Step 6 error "changing unwrap_gold_n_win does not work"):
- Run TopoPhaseremoval on ifg, then save lat and lon bands to gamma format, and rewrite in the geo folder (from: Step 6 error "changing unwrap_gold_n_win does not work"):
- Bigger subset area (50 km2 -> 200 km2): still have the error, not 10 but 19. After that i try with 400 km2, still get an error. Now i create the 1200 km2 subsets…
- Set the ‘unwrap_gold_n_win’ parameter from 32 to 18 (from MAINSAR)
- Set the ‘unwrap_grid_size’ parameter from 200 to 150 (from MAINSAR) -> It works, but maybe thats a wrong way or a dead end.
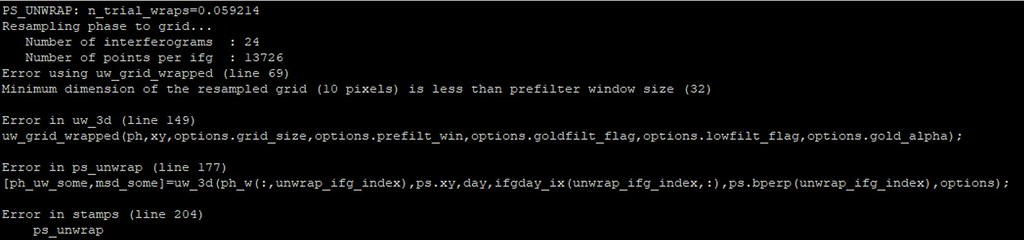
Now i haven’t get that error, but i had another:
Any suggestion will be great 
Regards, Andrew
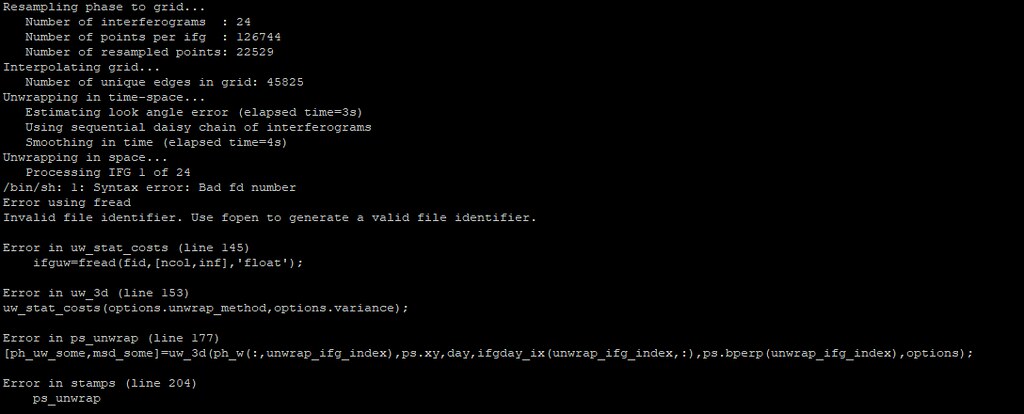
Not sure, but seeing the error is produced by the fread, my first thought comes on snaphu, do you have it installed it on the system? Can be called from MATLAB?
Please check whether from MATLAB you can call snaphu to be sure the command is found.
I hope this helps.
Let me know
Hi, thanks for your reply!
Yes, i have installed the snaphu, and set the PATH with:
PATH=“${PATH}:/stamps/snaphu-v1.4.2_linux/bin:/stamps/triangle:/stamps/StaMPS_v3.3b1/bin:/stamps/StaMPS_v3.3b1/matlab:/opt/matlab/R2016a/bin”
Now i did it with:
export SNAPHU_BIN=/stamps/snaphu-v1.4.2_linux/bin
and i try the whole process with the same subset area.
I used snaphu before, without the StaMPS+Matlab. With StaMPS, the step 6 call the snaphu automatically through the SNAPHU_BIN variable am i correct? How can i check, that the matlab can call the snaphu component corretly? I have tried the snaphu command in the matlab console, did not work (and i didn’t find and example to test that).
Thank you!
if SNAPHU_BIN is correctly entered in StaMPS_CONGIG (tcsh or bash) and you source it before starting matlab (from exactly this shell), it should be callable from within matlab.
Hi, the SNAPHU_BIN is correct in the config, and i source it before start the matlab. I got the “Minimum dimension of the resampled grid (26) is less than prefilter window size (32)” error, like before. So i had set the unwrap_grid_size parameter from 200 to 150 (tip from MAINSAR) to move on (maybe that a dead end solution). Now the new error is:
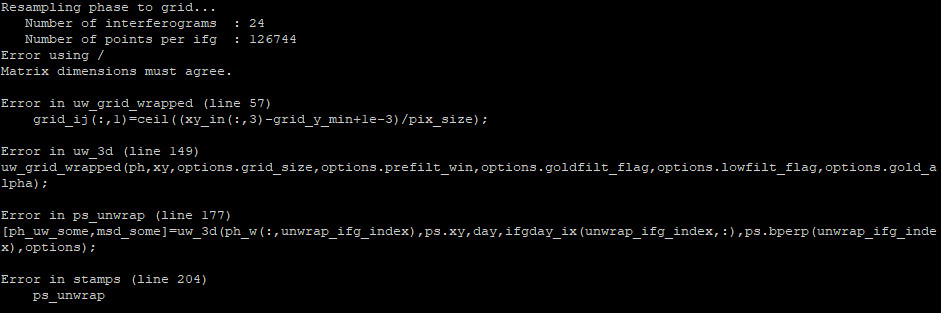
Now i see, twburns got the same error when he had run stamps step 6. I will try to run from the beginning (mt_prep…), maybe it will works fine.
Thank you!
Andrew
It is solved automatically, when i used bigger study area (ca. 800 km2), but i got a new matlab error at the second stamps step, which can be solved by following this: https://uk.mathworks.com/matlabcentral/answers/15521-matlab-function-save-and-v7-3
I had to edit these files:
- ps_load_initial_gamma.m
- ps_est_gamma_quick.m
Andrew
Hi,
I get the same error:
Error using uw_grid_wrapped (line 84)
Minimum dimension of the resampled grid (30 pixels) is less than prefilter window size (32)
Error in uw_3d (line 155)
uw_grid_wrapped(ph,xy,options.grid_size,options.prefilt_win,options.goldfilt_flag,options.lowfilt_flag,options.gold_alpha,options.ph_uw_predef);
Error in ps_unwrap (line 235)
[ph_uw_some,msd_some]=uw_3d(ph_w(:,unwrap_ifg_index),ps.xy,day,ifgday_ix(unwrap_ifg_index,:),ps.bperp(unwrap_ifg_index),options);
Error in stamps (line 503)
ps_unwrap
In according to MAINSAR group (https://groups.google.com/forum/#!topic/mainsar/5zTFtQb9OeM) I set the value of unwrap_gold_n_win to 30 with setparm('unwrap_gold_n_win',30). Then I ran the stamps(6,6) command.
And step 6 is completed and I didn’t take any error message.
Now, can this way be the solution to the problem? Or, should I follow the steps in the post marked as solution.
Thank you.
Hi dear. I got the same problem, can I ask you how did you can solve it??
thanks regard
Hello. I’m having the same problem.
How do I create the lat. and lon. file from the bands math? I’m working with COSMO SKyMed products, and I only do the coregistration and interferogram formation for the pre- process.
Hello,
You need to use bigger size area. Then try again.
Fikret
I am first time use the SNAP and stamps ,I meet the same error as yours, you say using Band Maths create the lat and lon bands, could you tell me how to do it? thanks you very much.
this is no longer required when you include the generation of the lat/lon and dem bands directly in the interferogram creation operator.
In the interferogram Formation I include the generation lat/lon like this,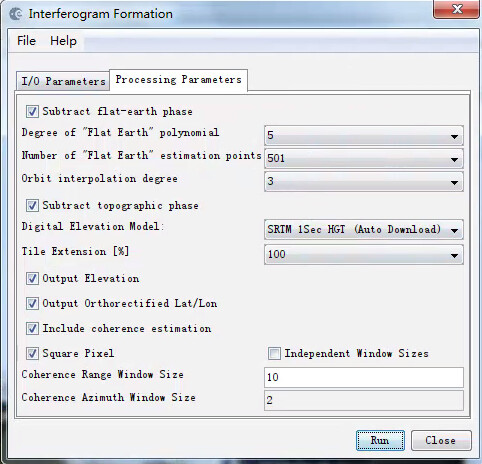
when I run the stamps(1,8); in MATLAB,I meet this error,
I saw the twburns’ answer about how to sovle this error,
But I am new to PS-INSAR,I don’t know how to use Band Maths to create lat and lon bands ? thanks for your reply.
this is a quite outdated solution and refers to the mt_prep_gamma_snap script which is no longer used.
So if the StaMPS export created the lat lon files and you ran mt_prep_snap successfully, I don’t think it will help to re-create the lat/lon bands manually.
But if you want to give it a try, you can replace the files in the DEM folder as suggested here: How to prepare Sentinel-1 images stack for PSI/SBAS in SNAP (step 9).
I rather suspect that snaphu might not be running correctly. What happens when you type snaphu in the command shell (not in matlab)?
i have the same error in the step 6. But i dont understand why this happend, i already run again all the process and the result is the same. Any idea how to fix this error?
was snaphu installed correctly?
yes, now i can run everything but i have one final problem, StaMPS complete sucesfull but when i call
the first command line ps_plot(‘v-dao’, ‘ts’) appears and error:
StaMPS returned an error “Unable to read file ‘./tca2’: no such file or directory.”
unable to load(apsname).
Days ago i haved the same problem and the solutions was reinstall StaMPS but now isnt work. Maybe i need change the StaMPS folder but i dont know how this can be a solution. i already modified snaphu and triangle folders.