Dear falahfakhri,
Thank you for your hard work in summerizing everything about using SNAP for PSI! It will be very helpful!
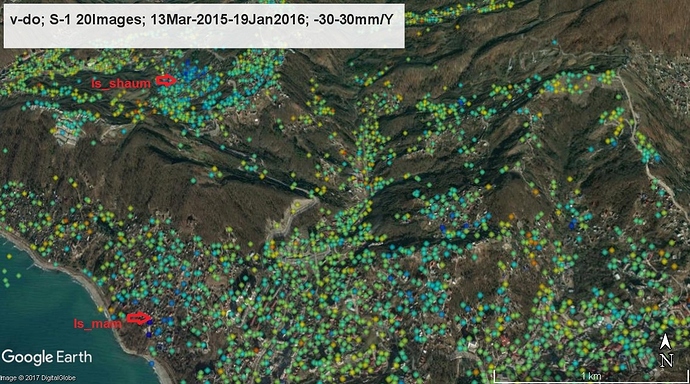
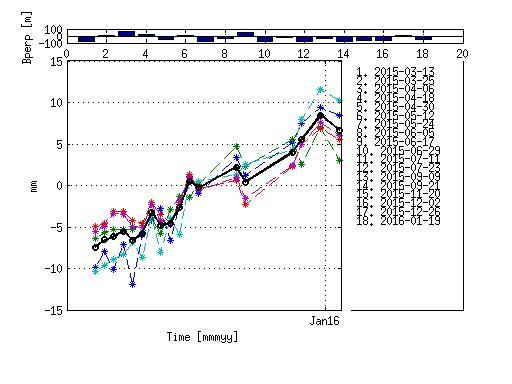
I am happy to share with my first reliable results obtained using Workflow between SNAP and StaMPS. Using 20 S-1 images I managed to fix the landslides which were previously fixed by processing of TSX, Envisat and ALOS data. Even ts graphs are not bad! The landslides are also fixed by ground data.
So it works!
#######################################################
Warning
The workflow described in this post (and the ones referring to it later on) is outdated and no longer recommended, because new versions of SNAP and StaMPS have been released. Please have a look at this updated list of instructions: StaMPS - Detailled instructions
#######################################################
I am glad to share my latest experience to save time for people doing PS processing of S-1 images using SNAP. Below is the short updated discription how I got these results.
snap-stamps
Tips. Use SNAP 6.0. beta (Earlier versions have bugs in Topophase removal). Visually inspect results of each step to be sure everything is OK. Try a small subset first.
1/ S-1 images: Split and Apply_Orbit_File. Split is done individually for each image because number of bursts covering the study area vary from image to image and you have to adjust this procedure manually. No batch processing can be used at this step. Apply orbit can be done using batch processing.
2/Create stack using Backgeocoding ( Radar-Coregistration-S-1Tops_Coregistration-S-1Backgeocoding. Master should be the first in the ProductSetReader). 12-13 images are OK for each Stack. If more images in the Stack - it is better to split it into several Stacks each having the same master. Master can be chosen by Radar-Interferometric-Insar Stack Overview.
3/Deburst the result of 2/
4/ Make subset of the result of 3/ (geographical coordinates) (Raster-Subset). Result file: subset_master_Stack_Deb.
5/ Apply Interferogram formation to the result of 4/ -. Result file: subset_master_Stack_Deb_ifg¬.
6/ Apply TopoPhaseRemoval to the result of 5/ . Result file: subset_master_Stack_Deb_ifg_dinsar.
7/ Add elevation band to the result of 6/ : subset_master_Stack_Deb_ifg¬_dinsar .
8/ Do Terrain Correction (TC) of the two products: subset_master_Stack_Deb and subset_master_Stack_Deb_ifg¬_dinsar
check boxes : output complex data and Latitude&longitude (when doing TC of subset_master_Stack_Deb). Without TC results of Stamps may be shifted in a strange way.
9/ Export data for StaMPS: Radar-Interferometric-Psi/sbas-StaMPS export
I exported to StaMPS the two files: subset_master_Stack_Deb_ifg_dinsar_TC and subset_master_Stack_Deb_TC . After the Stamps Export procedure I got four directories /diff0, /geo,/dem,/rslc which I placed into the directory INSAR_master_data. In case of several Stacks export each Stack and then copy results to the directories directories /diff0, /geo,/dem,/rslc.
10/ To have PS geocoded export subset_master_Stack_Deb_TC (with latitude and longitude bands) to ENVI or GAMMA format. Save result, for example, to /geo directory. Rename files with lon and lat to master_data.lon and master_data.lat. These are binary files with latitudes and longitudes for all pixels of the master crop. (One file contains values of lat for all pixels and the other - of lon) All the other files (results of export of the other bands) can be deleted. The files master_data.lon and master_data.lat should be placed into /geo directory.
You can also add longitude and latitude bands to subset_master_Stack_Deb, then do TC and get subset_master_Stack_Deb_TC_lon_lat. Export this to ENVI or GAMMA format.
Then like in 10/
There is a bug when exporting to Stamps TC results. You always get wrong value of heading in masterdata.rslc.par (it is always equal to 180deg) and thus in StaMPS. The easiest way to overcome this is to change “heading” in /rslc/masterdata.rslc.par manually to a correct value.
10/ Place the new scripts mt_prep_gamma_fei (changed by FeiLiu) to the /bin directory of StaMPS and ps_load_initial_gamma (changed by FeiLiu).m to the StaMPS matlab directory. ps_load_initial_gamma(changed).m (5.8 KB) please do no longer use these scripts, StaMPS was officially updated
mt_prep_gamma_snap(changed) (6.4 KB) please do no longer use these scripts, StaMPS was officially updated
Rename them into mt_prep_gamma and ps_load_initial_gamma.m correspondingly.
11/ run mt_prep_gamma from the INSAR_masterdata directory. mt_prep_gamma masterdata /fullpath to the INSAR_masterdata directory 0.4
12/ run matlab and launch StaMPS.
Try first step of stamps (1,1) - to be sure that all data are imported properly. Then continue.
No principal changes in Stamps parameters should be done. Even with default values I managed to get reliable results. Although to improve results adjustment of Stamps parameters is certainly necessary.
Good luck!
Many thanks to the developers and people who shared their experience in adjusting this soft!
How is your work on SBAS export? I will be glad if I can help. I am not an expert in writing scripts but I can test.
All the best,
Katherine