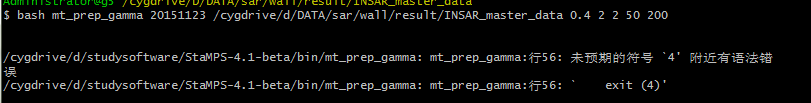
can you gudie me to pass the first step,mt_prep_gamma
all i have done is in the SNAP,so in the .bash file I dont know which to edit!
if you edit StaMPS_CONFIG.bash and execute it every time before you are going to use StaMPS, you don’t need to modify the system’s bash. You can do either the first OR the second option - both make the StaMPS scripts available to your shell.
another problem,the I download the triangle.zip,and do i need othe process to triangle(have unzipped)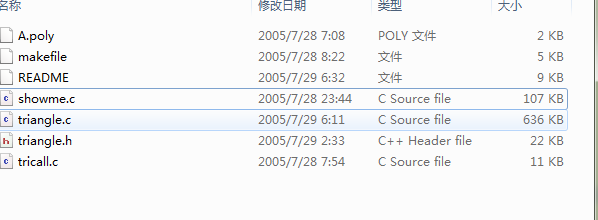
You need to compile triangle before you can use it. This is all documented well in the README file.
ok,i will try it
Just some remarks:
-
If you uses SNAP as InSAR processor, you can avoid to do modifications on the StaMPS_CONFIG.bash for triangle and snaphu
-
You can run StaMPS steps 1 to 5 without triangle and snaphu using maltab, by adding the stamps/matlab folder to the current path in matlab by:
addpath(’/path_to_stamps/matlab/’,path) -
Please ensure you uses the latest version of stamps (not released but the latest from github) to run at least step 1, as it solves many problems when SNAP writes 0’s or NaN’s pixels, while for the latest steps and plotting I suggest you to use the official release
Enjoy!
Dear all
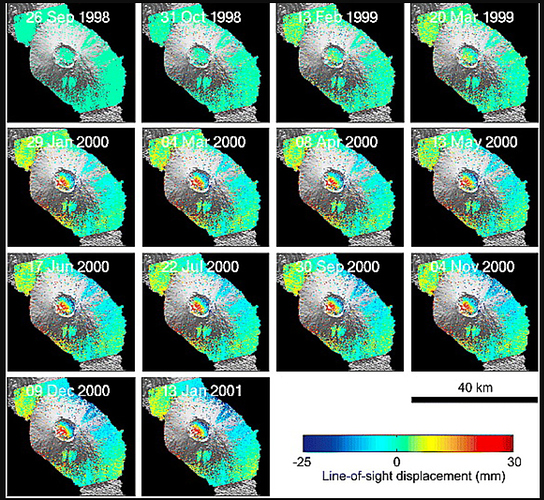
Using the ps_plot(‘u’) command, I can only obtain the unwrapped difference phase in time series.
I just want to get LOS displacement just like this figure in Andy’s PhD paper:
Which command I can use to get the serial LOS displacement ??
ps. The method I known: the unwrapped difference phase wavelength firstly, and then divide 4pi.
I am truly appreciate and welcome any kind help or advice.
JAMIE
Hi @katherine. The summary of the procedure was really helpful. I was able to succesfully export to StaMPS where I have the 4 folders (dem ,diff0, geo, rslc). I just can’t find a step by step procedure on how to proceed after the successful StaMPS export. Could you provide a document stating the steps on how to use StaMPS? Thank you.
you can follow the StaMPS tutorial, starting with chapter 6
https://homepages.see.leeds.ac.uk/~earahoo/stamps/StaMPS_Manual_v3.1.pdf
Thanks @ABraun. This is the document that I’m using but I can’t seem to follow the procedure. For instance, I don’t know what to with the first step. I can’t also produce an output when using mt_prep_snap. I would like to thank the authors for providing a manual but I would like to know if there’s a more detailed version (document that shows what to do with specific errors).
take these, for example:
- http://www.isnet.org.pk/downloadables/Presentation%20%20No.%202%20-%20STAMPS%20Steps.pdf Start with Phase 3, all previous steps have been performed in SNAP. You also need to call mt_prep_gamma_snap instead
- http://seom.esa.int/landtraining2015/files/Day_4/D4P2a_LTC2015_Hooper.pdf This document explains a bit more, what the single steps mean
- http://www.insar.cz/diplomky/diz_lazecky.pdf Chapter 3.3.2
If you can describe the errors you get, we can maybe help
Hello and happy to be among you.
please I would like to know if it is possible to realize all the steps of PSI Stamps in a windows operating system without involving the Linux system?
cordially
Thank you for reply please?
because i’m stuck on a step and i’m working on windows 10.
Hi @ABraun. I’ll take a look at these documents. I think it will be a huge help. Thank you!
Dear friends
Please help me to do PSI in SNAP-StaMPS and R-Studio.
I am preparing a stack of 33 S-1A ascending products to export using SNAP Stamps export. Following:
How to prepare Sentinel-1 images stack for PSI/SBAS in SNAP 5
Then i have to use R- studio.
Please send me list of commands and other resources to complete it through R studio.
StaMPS is written for Linux command line and Matlab. As far as I know there is no R version of it.
But you can use Python to prepare your files in a consistent and correct way. Everything is explained here: Snap2stamps package: a free tool to automate the SNAP-StaMPS Workflow
But after preparation of the SAR files, you will have to switch to Linux shell and Matlab.
ok i will try. But i have R studio installed on my Virtual Machine.
Don’t know how to run python there?
Just as Python, R studio is just a programming environment - you need scripts which define what is to do.
- snap2stamps uses Python to prepare Sentinel-1 SLC products for PS InSAR
- StaMPS uses Matlab to process the prepared products
None of the methods was developed for R so far.
ok thank you so much.