hi all
I am new to SAR, I am not a real programmer and possess little ego so any comment of the below workflow or advice on how to handle SAR data in a better fashion is much obliged.
I have taken a project which aims to detect soil erosion in a seasonally flooding river bed in a semi-arid environment. I was planning to compute interferometry for some 65 images and image combinations - you can imagine how many image pairs that is…
i have been looking at similar issues posted in Graph Builder for InSAR processing but hope there is newer suggestions for this issu
So this note is to those who have experience doing this sort of workflow….
There are two key issued here:
The first is some basic general questions about sar data and handling and the second – problems related to the execution of the a wanted workflow. Ill start with the latter.
Workflow:
I was hoping to get a workflow using the snap graphs (which I have built, see below for details) but it’s not working, not in unix and not in windows environment. However taking individual steps in windows I was able to run the entire process with good results (in unix – it failed this too). Running this process on individual pairs step by step is not realistic so I thought maybe with the combo of python+snappy I’ll have better luck in stringing the whole process together. If anyone has a working code – I would love to test –modify for myself.
To save time I’ll just describe the process I want to execute (I can send the graph itself for your testing upon request) but for now I’d like to have some commens on how to build this in python snappy
The steps are
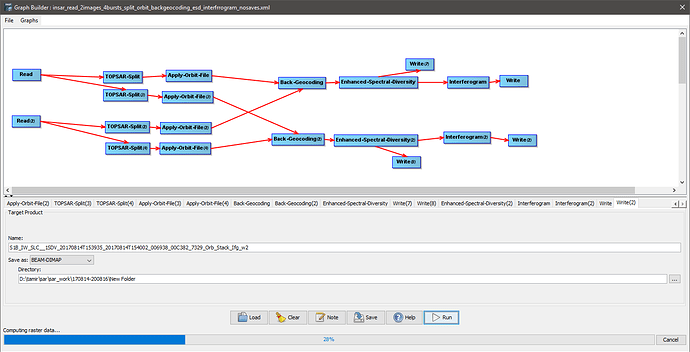
Read images (2img) >topsarsplit(2img)*>apply orbit file(2img) >>(and both into)>> back geocoding>enhanced spectral diversity filter (doesn’t work in a graph but does individually)>interferogram>topsar deburst>topo phase removal>goldstein phase filtring>write (ready for snaphu outside snap)
I made a bunch of scripts (graphs/models) in snap (v8) but it doesn’t work
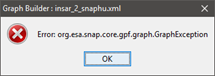
The first problem is I get the following error when I first try to open the graph for work
So
I tried to split it into sections and found that I can run the first 3 steps (topsarsplit(2img)>apply orbit file(2img) >>(and both into)>> back geocoding) but it gets stuck after that (the enhanced spectral diversity op. doesn’t work). I tried removing that step and I managed to run everything else…
Can anyone suggest why it doesn’t work etc etc ? Is this step necessary? are there any alternatives?
And to a more general question:
In the splitting op (topsar-split) I am asked to select the subswath (w1 /w2/w3) and the burst. My study site is spared on both w1 and w2 and on all 1-9 bursts for both swats. There is no option in the splitting step to include 2 swaths, is there? If I an use only one at a time (and go through each swath separately –that is I split the image into two - w1 and w2) - at what point can I merge them together into a single output?
Ill be very glad for some help and ill update here if and when I figure out solutions
But please if you have ideas for a completely different approach – one I didn’t think about – please suggest.
Many thanks in advance
Tamir
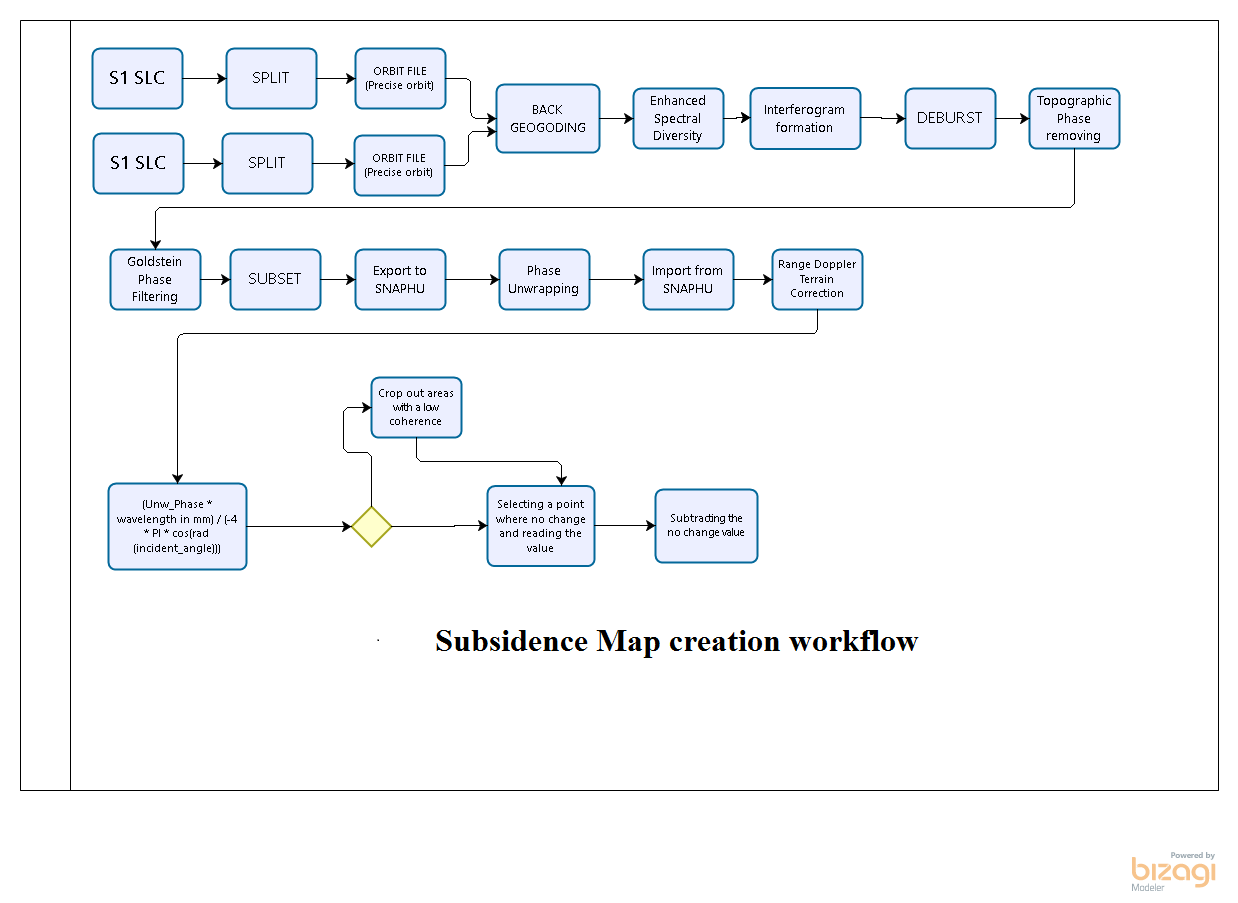
 i have done exactly as you suggested (great minds think alike). and to that end - i already followed both your advice and separated run all ops. everything work well and results are good (the errors i has were only when trying to graph them together).
i have done exactly as you suggested (great minds think alike). and to that end - i already followed both your advice and separated run all ops. everything work well and results are good (the errors i has were only when trying to graph them together).