i install python2.7 in ubuntu18.04 and problem has been solved
Not sure if this has been posted elsewhere already, but in case it helps, even with SNAP 8 (Ubuntu 18.04) I had to change those lines in all three coreg_ifg xml files to resolve Operator ‘SpectralDiversityOp’: Unknown element ‘useSuppliedShifts’ error.
Hello everyone,
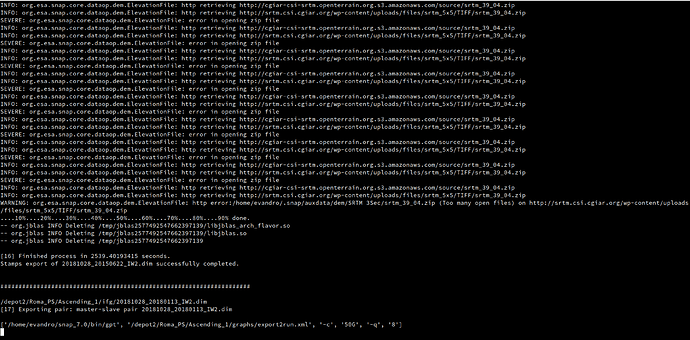
I’m facing an issue during the export step of snap2stamps. As you can see in the image below, the export takes really a lot of time, ca. 40 min each (for comparison, the coregistration took 5/7 min each) while usually with the same computing resources emploied, it takes a few minutes. Furthermore, despite the export is marked as “succesfully completed”, there are several “severe” and a “warning” whose meaning is not clear to me. I fear that the exports will be somoheow “corrupted”.
I’m running this step with SNAP 7.0, I also tried with SNAP 8.0 but the folder INSAR_… was incomplete. In fact, using SNAP 8.0 the entire “dem” folder and the .diff and .base files for each export within the “diff0” folder were missing.
Does anyone have an idea about this issue or how to solve it?
Thank you in advance!
I feel it is due to a network issue.
Stamps export will take more time compared to the remaining steps.
Is there any error, while processing the coregistration and Enhanced spectral diversity step?
No, everything went well. I also cheked each inteferogram one by one to be sure they were created correctly. For network issue, do you mean some problem related to SNAP/Copernicus servers?
- For determining the lonmin, latmin, lonmax, latmax values for AOI use this below website.
Instead of MARC format just select the CSV format, then directly copy those lat long to your project.conf file.
- Whatever area covering by master image (6 bursts) only will process. you will get result only master image covering area.
Maybe the master image covers only that particular portion (only 6 bursts). Better you just change the master (Delete the old master image and try to incorporate another master image).
Better you just try to use Python script (snap2stamps). It is very easy to process.
GitHub - mdelgadoblasco/snap2stamps: Using SNAP as InSAR processor for StaMPS
Hello everyone,
I’m having some problem with the coregistration and the stamps export steps
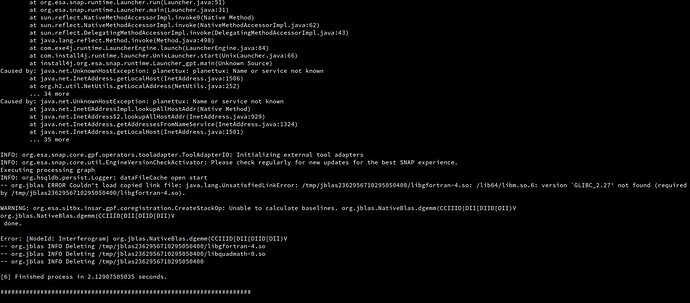
- Coregistration issue: using the most recent updated version of SNAP 8 it fails giving the error reported in the image below. I saw in other answers in the forum that the suggested solution is to download libgfortran3 or 5 but I already have both installed and related to a GCC v 10.3. I also tried to re-install them but the problem persists.
- Stamps export issue: Coregistration does not work with SNAP 8 but it does with SNAP 7, so I proceeded. Unfortunately the export has problems as well with both SNAP versions.
- Using SNAP 8, the INSAR… folder was incomplete: the entire “dem” folder was missing as well as the .diff and .base files for each export within the “diff0” folder.
- Then I tried with SNAP 7, the process starts but it is endless…
Can someone help me?
Thank you very much!
Try this below one. Maybe it will resolve your problem.
Does this work also with OpenSuse?
yes. It will work.
Thank you both, “unfortunately” I already have gcc-7 selected as preferred compiler
Can you show us the output from which gcc and gcc --version (sometimes a gcc provided by a third party gets priority by position in the PATH variable)? If you have object (.o) or library files create with the wrong compiler you should delete them so they can be regenerated with the correct compiler.
I updated OpenSuse from v15.2 to 15.3 and now both coregistration and export work! I checked one by one each interferogram and they are ok and the export creates all the four folders and all the expected files within.
Maybe the problem was with the GLIBC.
OpenSuse v15.2 has GLIBC_2.26 while OpenSuse v15.3 has GLIBC_2.31
Finger crossed for the next steps!
Glad you found the solution. It is somewhat rare to get such a clear error message as /lib64/libm.so.6, version GLIBC_N.N.N not found. The linux glibc is actually well designed
and incorporates years of in-service experience.
Software built on a system with a newer glibc rarely, if ever, will run on older glibc versions. All the effort goes to making sure that software built with an older glibc will still run on a newer version.
PS – your screen image difficult to read on a small laptop screen, and is not searchable, so won’t be found by the next person who gets the same error – posting such messages as plain text helps build
a long-term archive of searchable problems and solutions.
Maybe one or multiple corrupted files somhow were disturbing the processing and the software. Thank you for the suggestion, I post here below the text of the error:
– org.jblas ERROR Couldn’t load copied link files: java.lang.UnsatisfiedLinkError: /tmp/jblas2362956710295050400/libfortran-4.so: /lib64/libm.so.6: version ‘GLIBC_2.27’ not found (required by /tmp/jblas2362956710295050400/libfortran-4.so).
While preprocessing sentinel-1 data for StaMPS I got “Error: [NodeId: Back-Geocoding] Split product is expected.”. The provided master and slave images both split products. If I run this process manually it works fine. However, if I do it using snap2stamps it give the following error. Any hint would be appriciated.
(base) mirza@lumir:~/InSAR/Korea/Busan/2016/des/Project/bin$ python2 coreg_ifg_topsar.py project.conf
/home/mirza/InSAR/Korea/Busan/2016/des/Project
/home/mirza/InSAR/Korea/Busan/2016/des/Project/graphs
IW2
/home/mirza/InSAR/Korea/Busan/2016/des/Project/master/20160718_IW2.dim
/opt/snap/bin/gpt
/home/mirza/InSAR/Korea/Busan/2016/des/Project/graphs/coreg_ifg_computation_subset.xml
#####################################################################
Coregistration and Interferogram computation started:
#####################################################################
/home/mirza/InSAR/Korea/Busan/2016/des/Project/split/20160308/20160308_IW2.dim
[1] Processing slave file :20160308_IW2.dim
SNAP STDOUT:INFO: org.esa.snap.core.gpf.operators.tooladapter.ToolAdapterIO: Initializing external tool adapters
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: GDAL not found on system. Internal GDAL 3.0.0 from distribution will be used. (f1)
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.
INFO: org.esa.snap.core.util.EngineVersionCheckActivator: Please check regularly for new updates for the best SNAP experience.
INFO: org.esa.s2tbx.dataio.gdal.GDALVersion: Internal GDAL 3.0.0 set to be used by SNAP.
Executing processing graph
INFO: org.hsqldb.persist.Logger: dataFileCache open start
Split product is expected.
done.
Error: [NodeId: Back-Geocoding] Split product is expected.
[1] Finished process in 2.8753221035 seconds.
Try this below one:
I already did this but still getting the same error.