yes, please have a look at this post: StaMPS - Detailed instructions
Hi Gijs,
Could you resolve this issue? I’m having the same problem…
Sohrab
Check this below one, For me it’s working.
Thanks a lot suribabu! but unfortunately that didn’t work for me…
Apparently, not so many people had the same issue so probably that’s something related to the dataset.
hey, i ran into the same problem. have you found the solution?
Remove inverted commas and run
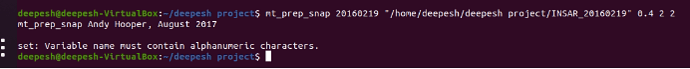
The space in a directory name likely causes your problem.
instead of INSAR_20160219 just give INSAR_20160219/
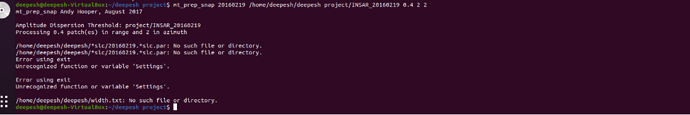
i tried with both suggestions, it worked but another error came
is it my system? i downloaded ubuntu in virtualbox and set up everything for stamps
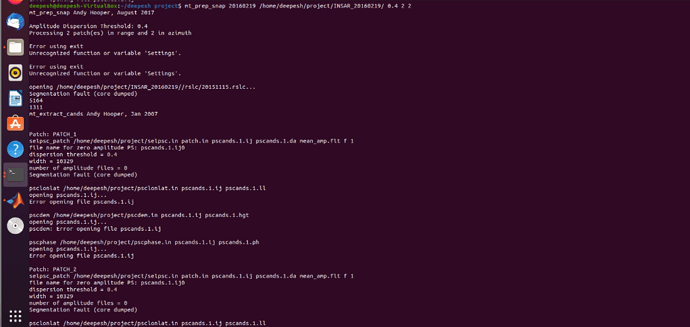
The “segmentation fault (core dumped)” error occurs because StaMPS was compiled with the wrong compiler. You find a solution here: Segmentation Fault (core dumped) error during mt_prep_snap - #17 by ABraun
In my case this error was caused by failing to start a new project with a fresh INSAR_XXXXXXXX directory. Solved by clearing old files from the directory then rerunning mt_prep_snap.
Hello Everyone,
I have a simple question regarding geocoding process.
In SNAP-StaMPS processing steps, in which step the geocoding is done?
I have checked one of xxxxx.rslc.par file in PSINSAR_20180425/rslc folder, it says “image_geometry: SLANT_RANGE”.
It means that the image is still in range and azimuth (radar coordinate) after “7ps_export.py”.
However, in matlab, when I run "ps_plot ‘u’ " after step-6 (StaMPS) the unwrapped phases is already in longitude and latitude coordinate.
So, is anyone know in which step the geocoding was done?
Thank you.
the variables are explained in the script
Dear all,
I have got a PSInSAR result by applying snap2stamps package to process 177 images from 2014/10 to 2019/10.
Now, I want to analyze the time series after 2019/10.
Do you suggest to acquire the time series of surface deformation more than 6 years by PSInSAR method? I think it would be too long for PSInSAR analysis…
Or I could only process the images after 2019/6 (maybe a short overlap period with the previous result which end at 2019/10), and then connected these two time series? But they are with different master images and the distribution of PS pixels may be different. The time series may be acquired by calculated the mean displacement of PS pixels in a small region of the results based on two different time period.
Or…I need to apply the SBAS method to deal with this problem?
Hope someone could give me some suggestions. Thank you very much!
Hi @bayzidul I was wondering if your first picture represents StaMPS successfully installed , Since my matlab cannot call stamps, I can’t judge whether I successfully installed stamps or whether there are problems in other links. We look forward to your reply
Have you set the path of StaMPS in your Matlab?
Sorry @suribabu First of all, does that picture represent a successful installation? Second, the tutorial I used didn’t teach me how to set StaMPS paths in MATLAB. Can you tell me how to do that,thank you very much.
First of all, Click on ‘setpath’ in Matlab (window shows like below picture), In setpath window click on Add with subfolders… then select the StaMPS folder (the downloaded StaMPS path).
Once you will add the path, just run stamps(1,1) command in matlab.
But before running the stamps in Matlab, you should complete the mt_prep_snap step in terminal.